Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6413
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7561 | 3.375e-03 | 5.430e-04 | 7.647e-01 | 1.401e-06 |
|---|
| Loi | 0.2296 | 8.144e-02 | 1.167e-02 | 4.311e-01 | 4.097e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.574e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.500e-03 | 0.000e+00 |
|---|
| Wang | 0.2606 | 2.810e-03 | 4.501e-02 | 4.019e-01 | 5.083e-05 |
|---|
Expression data for subnetwork 6413 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
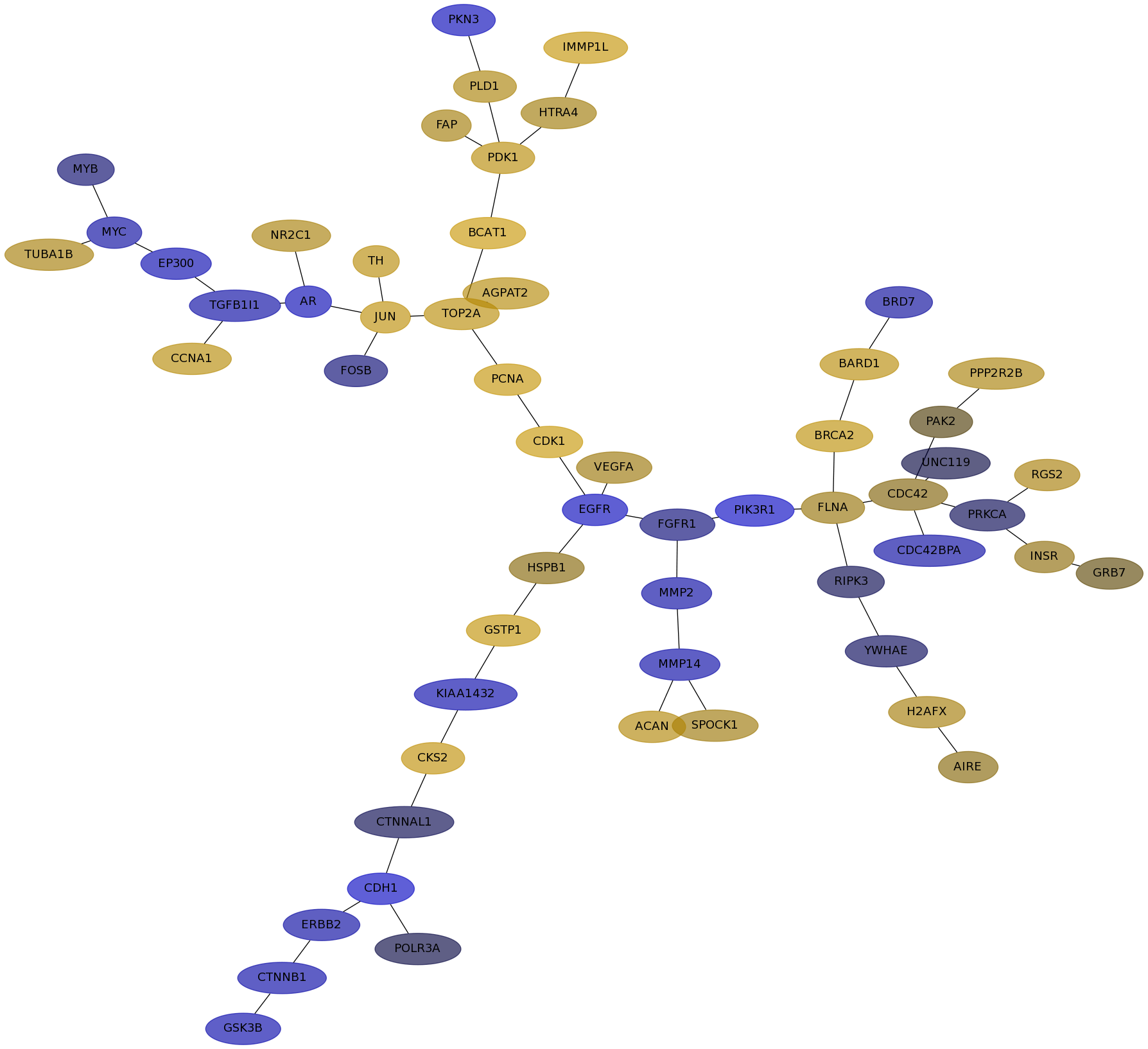
Score for each gene in subnetwork 6413 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SPOCK1 |   | 1 | 1195 | 2087 | 2092 | 0.093 | 0.253 | 0.145 | 0.207 | 0.241 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| BCAT1 |   | 4 | 440 | 1735 | 1699 | 0.273 | -0.134 | 0.161 | -0.078 | 0.122 |
|---|
| RGS2 |   | 3 | 557 | 1131 | 1126 | 0.118 | 0.061 | 0.084 | 0.050 | 0.178 |
|---|
| YWHAE |   | 4 | 440 | 179 | 209 | -0.018 | 0.112 | 0.279 | -0.094 | 0.014 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| KIAA1432 |   | 3 | 557 | 1681 | 1657 | -0.132 | -0.054 | undef | 0.108 | undef |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| MMP2 |   | 3 | 557 | 1190 | 1182 | -0.105 | 0.186 | -0.024 | undef | 0.162 |
|---|
| FOSB |   | 5 | 360 | 1534 | 1501 | -0.033 | 0.046 | 0.129 | -0.104 | 0.095 |
|---|
| MMP14 |   | 2 | 743 | 1190 | 1189 | -0.114 | 0.056 | 0.011 | 0.105 | 0.148 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| FAP |   | 1 | 1195 | 2087 | 2092 | 0.110 | 0.065 | -0.011 | 0.122 | 0.181 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| CTNNAL1 |   | 4 | 440 | 682 | 678 | -0.014 | 0.073 | -0.008 | -0.131 | 0.004 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| CDC42BPA |   | 6 | 301 | 1268 | 1236 | -0.102 | 0.098 | 0.059 | 0.368 | 0.260 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| TUBA1B |   | 6 | 301 | 590 | 582 | 0.116 | 0.094 | 0.249 | 0.101 | 0.128 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| AGPAT2 |   | 3 | 557 | 412 | 430 | 0.167 | -0.015 | 0.123 | 0.042 | -0.076 |
|---|
| AIRE |   | 1 | 1195 | 2087 | 2092 | 0.054 | -0.163 | -0.033 | 0.097 | -0.196 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| ACAN |   | 5 | 360 | 412 | 416 | 0.156 | 0.132 | -0.024 | -0.171 | -0.083 |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| H2AFX |   | 7 | 256 | 179 | 201 | 0.111 | -0.072 | 0.121 | 0.165 | -0.010 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| TH |   | 2 | 743 | 1988 | 1970 | 0.182 | -0.018 | 0.068 | undef | 0.039 |
|---|
| IMMP1L |   | 2 | 743 | 1830 | 1815 | 0.243 | -0.028 | undef | undef | undef |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
GO Enrichment output for subnetwork 6413 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.744E-09 | 9.147E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.736E-08 | 2.121E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.116E-08 | 2.537E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.037E-08 | 3.687E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.109E-08 | 3.962E-05 |
|---|
| nucleus localization | GO:0051647 |  | 8.109E-08 | 3.302E-05 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 8.962E-08 | 3.128E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.213E-07 | 3.703E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 2.437E-07 | 6.614E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.413E-07 | 1.811E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 9.663E-07 | 2.146E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 5.415E-09 | 1.303E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.51E-08 | 3.019E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.502E-08 | 3.611E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.477E-08 | 4.498E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.508E-08 | 4.575E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.171E-07 | 4.696E-05 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.41E-07 | 4.848E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.751E-07 | 5.265E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 3.516E-07 | 9.399E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.162E-06 | 2.795E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 1.638E-06 | 3.583E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.355E-09 | 1.462E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.407E-08 | 5.068E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.772E-08 | 6.726E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.572E-07 | 9.037E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.595E-07 | 1.654E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.078E-07 | 1.563E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 4.43E-07 | 1.455E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 8.753E-07 | 2.517E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.093E-06 | 5.35E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.148E-06 | 4.94E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.86E-06 | 5.98E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.844E-07 | 5.242E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.476E-07 | 3.203E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 4.284E-07 | 2.632E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.192E-07 | 2.392E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.86E-07 | 3.266E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 1.949E-06 | 5.985E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.949E-06 | 5.13E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 3.881E-06 | 8.941E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.992E-06 | 1.227E-03 |
|---|
| regulation of protein transport | GO:0051223 |  | 1.359E-05 | 2.505E-03 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 1.61E-05 | 2.697E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.355E-09 | 1.462E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.407E-08 | 5.068E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.772E-08 | 6.726E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.572E-07 | 9.037E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.595E-07 | 1.654E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.078E-07 | 1.563E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 4.43E-07 | 1.455E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 8.753E-07 | 2.517E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.093E-06 | 5.35E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.148E-06 | 4.94E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.86E-06 | 5.98E-04 |
|---|



