Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6329
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7575 | 3.290e-03 | 5.250e-04 | 7.609e-01 | 1.314e-06 |
|---|
| Loi | 0.2291 | 8.212e-02 | 1.185e-02 | 4.327e-01 | 4.212e-04 |
|---|
| Schmidt | 0.6719 | 0.000e+00 | 0.000e+00 | 4.660e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.484e-03 | 0.000e+00 |
|---|
| Wang | 0.2602 | 2.868e-03 | 4.552e-02 | 4.042e-01 | 5.277e-05 |
|---|
Expression data for subnetwork 6329 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
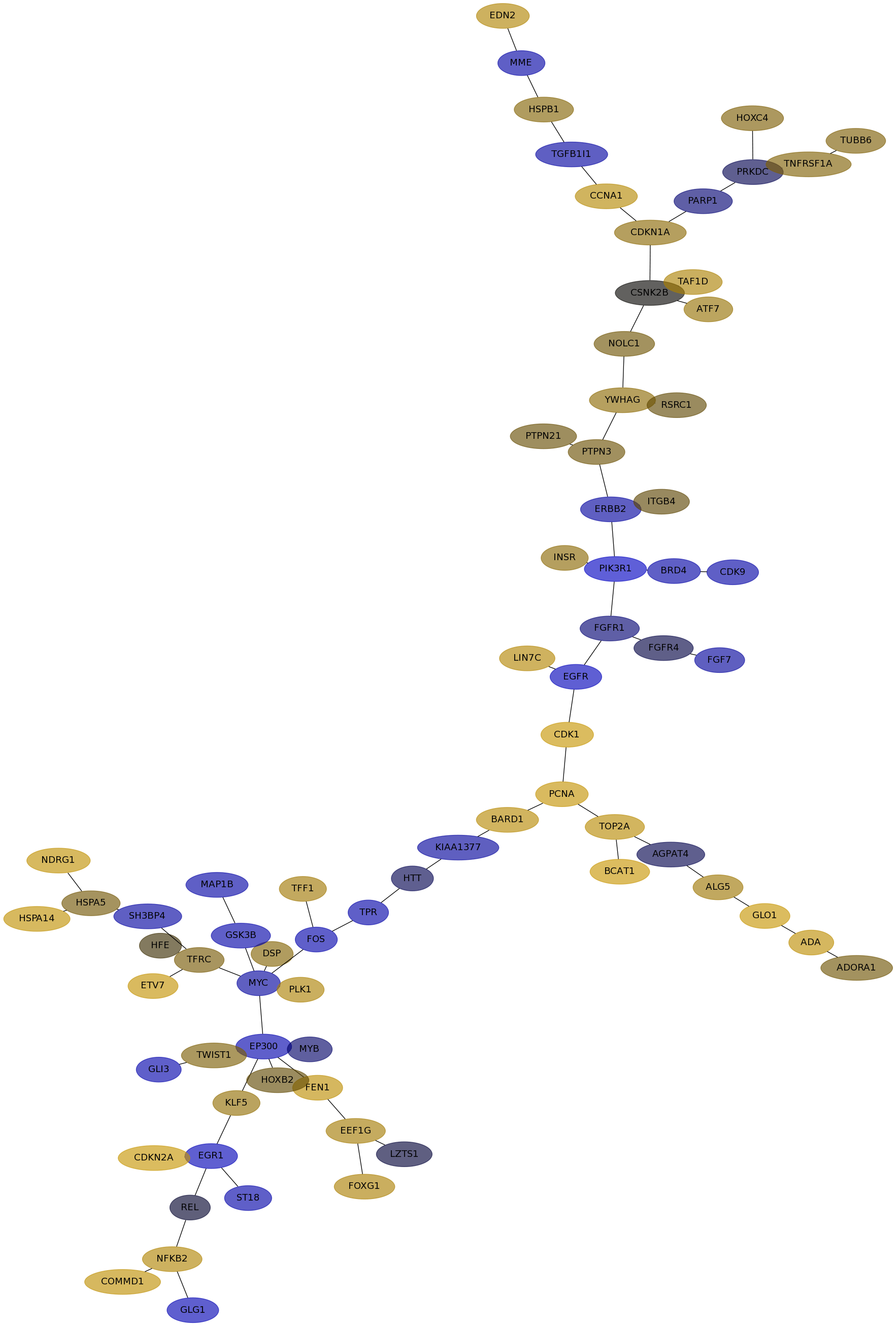
Score for each gene in subnetwork 6329 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NDRG1 |   | 6 | 301 | 658 | 646 | 0.222 | 0.143 | -0.040 | -0.018 | 0.242 |
|---|
| PTPN21 |   | 5 | 360 | 1717 | 1677 | 0.027 | 0.177 | 0.214 | 0.205 | 0.119 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| BCAT1 |   | 4 | 440 | 1735 | 1699 | 0.273 | -0.134 | 0.161 | -0.078 | 0.122 |
|---|
| CSNK2B |   | 3 | 557 | 318 | 341 | 0.001 | -0.039 | -0.004 | 0.115 | 0.033 |
|---|
| TUBB6 |   | 3 | 557 | 1461 | 1440 | 0.045 | 0.114 | 0.187 | undef | 0.165 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| FEN1 |   | 2 | 743 | 2024 | 2002 | 0.210 | -0.132 | 0.008 | 0.002 | 0.028 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| HSPA5 |   | 2 | 743 | 2180 | 2165 | 0.035 | 0.033 | -0.006 | undef | -0.203 |
|---|
| GLO1 |   | 5 | 360 | 83 | 110 | 0.266 | -0.089 | 0.135 | 0.100 | 0.076 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| HFE |   | 1 | 1195 | 2180 | 2186 | 0.009 | 0.062 | -0.133 | undef | 0.016 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| EDN2 |   | 3 | 557 | 1116 | 1107 | 0.153 | 0.062 | 0.132 | 0.196 | 0.287 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| TNFRSF1A |   | 8 | 222 | 412 | 413 | 0.050 | 0.094 | -0.014 | 0.214 | -0.045 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| NOLC1 |   | 4 | 440 | 1047 | 1032 | 0.033 | -0.055 | 0.172 | 0.012 | -0.064 |
|---|
| LZTS1 |   | 3 | 557 | 2147 | 2108 | -0.008 | -0.089 | 0.043 | 0.147 | 0.063 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| REL |   | 2 | 743 | 318 | 346 | -0.006 | -0.018 | -0.101 | -0.093 | -0.183 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| FOXG1 |   | 6 | 301 | 727 | 716 | 0.136 | -0.156 | -0.126 | -0.013 | -0.068 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| TFRC |   | 5 | 360 | 957 | 938 | 0.038 | -0.066 | 0.076 | -0.108 | -0.014 |
|---|
| ADA |   | 4 | 440 | 83 | 112 | 0.208 | -0.164 | -0.042 | -0.102 | 0.219 |
|---|
| HSPA14 |   | 1 | 1195 | 2180 | 2186 | 0.229 | -0.283 | -0.047 | -0.114 | 0.026 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| ADORA1 |   | 1 | 1195 | 2180 | 2186 | 0.033 | -0.079 | -0.119 | undef | -0.074 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| ETV7 |   | 4 | 440 | 957 | 942 | 0.245 | -0.201 | -0.043 | -0.062 | -0.174 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| AGPAT4 |   | 2 | 743 | 1 | 66 | -0.014 | 0.005 | 0.059 | -0.020 | 0.108 |
|---|
| PRKDC |   | 1 | 1195 | 2180 | 2186 | -0.015 | -0.132 | 0.147 | 0.209 | 0.057 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TFF1 |   | 7 | 256 | 957 | 928 | 0.106 | 0.022 | 0.088 | -0.033 | -0.244 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| COMMD1 |   | 1 | 1195 | 2180 | 2186 | 0.217 | -0.124 | 0.216 | -0.167 | undef |
|---|
| ALG5 |   | 1 | 1195 | 2180 | 2186 | 0.098 | -0.090 | -0.018 | 0.009 | 0.059 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| CDK9 |   | 3 | 557 | 1598 | 1574 | -0.120 | 0.179 | 0.182 | 0.186 | -0.135 |
|---|
| GLG1 |   | 2 | 743 | 1437 | 1433 | -0.165 | 0.090 | -0.094 | 0.050 | -0.091 |
|---|
| SH3BP4 |   | 4 | 440 | 957 | 942 | -0.105 | 0.108 | -0.100 | 0.246 | -0.003 |
|---|
| HOXC4 |   | 1 | 1195 | 2180 | 2186 | 0.046 | 0.169 | 0.088 | 0.139 | 0.247 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| MME |   | 5 | 360 | 1047 | 1021 | -0.121 | 0.020 | 0.088 | -0.002 | -0.130 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6329 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic organ development | GO:0048568 |  | 4.287E-08 | 1.047E-04 |
|---|
| embryonic organ morphogenesis | GO:0048562 |  | 5.157E-08 | 6.299E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.037E-07 | 4.102E-04 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 5.237E-07 | 3.198E-04 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.322E-06 | 6.461E-04 |
|---|
| immunoglobulin V(D)J recombination | GO:0033152 |  | 1.613E-06 | 6.567E-04 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 1.613E-06 | 5.629E-04 |
|---|
| protein destabilization | GO:0031648 |  | 3.213E-06 | 9.811E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.711E-06 | 1.007E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 3.973E-06 | 9.705E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.391E-06 | 1.197E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic organ development | GO:0048568 |  | 6.807E-08 | 1.638E-04 |
|---|
| embryonic organ morphogenesis | GO:0048562 |  | 7.622E-08 | 9.169E-05 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 5.846E-07 | 4.689E-04 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.406E-06 | 8.458E-04 |
|---|
| immunoglobulin V(D)J recombination | GO:0033152 |  | 2.047E-06 | 9.848E-04 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 2.047E-06 | 8.207E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 3.46E-06 | 1.189E-03 |
|---|
| protein destabilization | GO:0031648 |  | 4.075E-06 | 1.226E-03 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 5.427E-06 | 1.451E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.809E-06 | 1.398E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.101E-06 | 1.553E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anterior/posterior pattern formation | GO:0009952 |  | 1.031E-09 | 2.371E-06 |
|---|
| embryonic organ development | GO:0048568 |  | 2.136E-07 | 2.456E-04 |
|---|
| embryonic organ morphogenesis | GO:0048562 |  | 2.717E-07 | 2.083E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 4.346E-06 | 2.499E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 4.453E-06 | 2.048E-03 |
|---|
| protein destabilization | GO:0031648 |  | 4.453E-06 | 1.707E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 4.453E-06 | 1.463E-03 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 7.487E-06 | 2.153E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.856E-06 | 2.263E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.856E-06 | 2.037E-03 |
|---|
| somitogenesis | GO:0001756 |  | 9.569E-06 | 2.001E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.568E-06 | 2.89E-03 |
|---|
| anterior/posterior pattern formation | GO:0009952 |  | 2.963E-06 | 2.731E-03 |
|---|
| embryonic gut development | GO:0048566 |  | 5.33E-06 | 3.274E-03 |
|---|
| embryonic organ development | GO:0048568 |  | 9.301E-06 | 4.285E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.06E-05 | 3.906E-03 |
|---|
| pattern specification process | GO:0007389 |  | 1.075E-05 | 3.303E-03 |
|---|
| lung development | GO:0030324 |  | 1.162E-05 | 3.061E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 1.516E-05 | 3.493E-03 |
|---|
| respiratory system development | GO:0060541 |  | 1.516E-05 | 3.105E-03 |
|---|
| cranial nerve development | GO:0021545 |  | 1.844E-05 | 3.398E-03 |
|---|
| regionalization | GO:0003002 |  | 1.98E-05 | 3.318E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anterior/posterior pattern formation | GO:0009952 |  | 1.031E-09 | 2.371E-06 |
|---|
| embryonic organ development | GO:0048568 |  | 2.136E-07 | 2.456E-04 |
|---|
| embryonic organ morphogenesis | GO:0048562 |  | 2.717E-07 | 2.083E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 4.346E-06 | 2.499E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 4.453E-06 | 2.048E-03 |
|---|
| protein destabilization | GO:0031648 |  | 4.453E-06 | 1.707E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 4.453E-06 | 1.463E-03 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 7.487E-06 | 2.153E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.856E-06 | 2.263E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.856E-06 | 2.037E-03 |
|---|
| somitogenesis | GO:0001756 |  | 9.569E-06 | 2.001E-03 |
|---|



