Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6267
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7593 | 3.187e-03 | 5.040e-04 | 7.560e-01 | 1.214e-06 |
|---|
| Loi | 0.2288 | 8.265e-02 | 1.200e-02 | 4.340e-01 | 4.303e-04 |
|---|
| Schmidt | 0.6712 | 0.000e+00 | 0.000e+00 | 4.729e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.484e-03 | 0.000e+00 |
|---|
| Wang | 0.2597 | 2.936e-03 | 4.612e-02 | 4.069e-01 | 5.510e-05 |
|---|
Expression data for subnetwork 6267 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
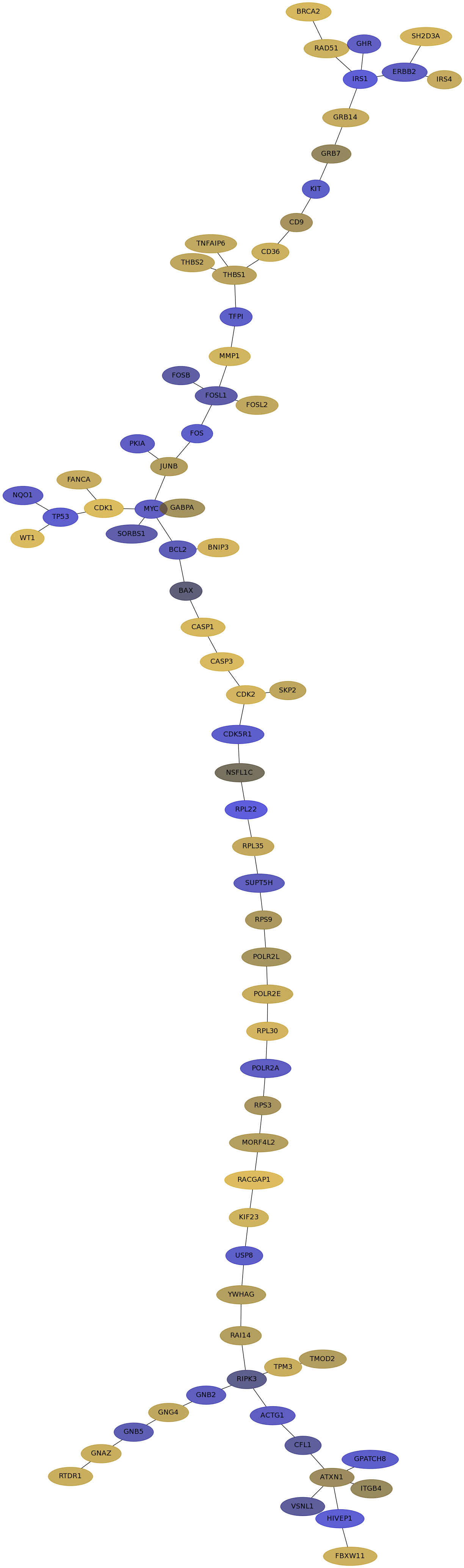
Score for each gene in subnetwork 6267 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| FBXW11 |   | 3 | 557 | 658 | 654 | 0.154 | 0.225 | 0.262 | undef | 0.169 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ACTG1 |   | 2 | 743 | 983 | 988 | -0.102 | 0.061 | 0.012 | 0.161 | 0.235 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| RPL30 |   | 1 | 1195 | 1903 | 1915 | 0.195 | -0.138 | 0.144 | -0.107 | -0.032 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| MORF4L2 |   | 1 | 1195 | 1903 | 1915 | 0.063 | 0.214 | 0.018 | 0.017 | 0.157 |
|---|
| FOSL2 |   | 3 | 557 | 366 | 389 | 0.100 | 0.080 | -0.119 | -0.154 | 0.080 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| FOSB |   | 5 | 360 | 1534 | 1501 | -0.033 | 0.046 | 0.129 | -0.104 | 0.095 |
|---|
| NSFL1C |   | 2 | 743 | 1576 | 1561 | 0.005 | 0.016 | -0.204 | 0.137 | -0.021 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| CFL1 |   | 2 | 743 | 1688 | 1673 | -0.027 | 0.087 | 0.010 | 0.303 | 0.008 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| POLR2L |   | 5 | 360 | 727 | 722 | 0.037 | -0.012 | 0.224 | 0.054 | 0.015 |
|---|
| TPM3 |   | 2 | 743 | 1903 | 1878 | 0.135 | 0.036 | undef | undef | undef |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| TFPI |   | 1 | 1195 | 1903 | 1915 | -0.129 | -0.004 | 0.087 | 0.044 | 0.157 |
|---|
| VSNL1 |   | 3 | 557 | 1437 | 1418 | -0.026 | 0.080 | 0.220 | -0.099 | -0.041 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| MMP1 |   | 11 | 148 | 366 | 360 | 0.192 | 0.014 | 0.036 | 0.044 | 0.260 |
|---|
| CD36 |   | 2 | 743 | 83 | 123 | 0.146 | 0.017 | 0.129 | 0.061 | 0.124 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| RPS9 |   | 1 | 1195 | 1903 | 1915 | 0.044 | -0.059 | 0.209 | -0.187 | -0.119 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| TMOD2 |   | 1 | 1195 | 1903 | 1915 | 0.061 | -0.081 | 0.055 | -0.061 | undef |
|---|
| RTDR1 |   | 5 | 360 | 1364 | 1328 | 0.135 | 0.098 | 0.222 | 0.125 | undef |
|---|
| RPL22 |   | 3 | 557 | 1576 | 1548 | -0.266 | 0.105 | 0.059 | -0.143 | 0.112 |
|---|
| RPS3 |   | 2 | 743 | 1903 | 1878 | 0.038 | 0.007 | 0.236 | -0.194 | 0.006 |
|---|
| FOSL1 |   | 11 | 148 | 366 | 360 | -0.040 | 0.164 | 0.010 | 0.128 | 0.005 |
|---|
| HIVEP1 |   | 6 | 301 | 764 | 758 | -0.199 | 0.069 | 0.000 | 0.193 | -0.042 |
|---|
| SH2D3A |   | 3 | 557 | 1510 | 1486 | 0.197 | 0.141 | -0.106 | -0.012 | 0.121 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| KIF23 |   | 4 | 440 | 957 | 942 | 0.168 | -0.063 | 0.162 | undef | 0.033 |
|---|
| RACGAP1 |   | 4 | 440 | 1222 | 1205 | 0.283 | -0.164 | 0.073 | 0.055 | 0.188 |
|---|
| THBS1 |   | 5 | 360 | 1141 | 1109 | 0.078 | 0.044 | 0.185 | 0.200 | 0.267 |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| BAX |   | 1 | 1195 | 1903 | 1915 | -0.006 | -0.068 | 0.107 | -0.255 | -0.110 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| CASP1 |   | 6 | 301 | 552 | 544 | 0.204 | -0.166 | -0.046 | undef | -0.034 |
|---|
| TNFAIP6 |   | 3 | 557 | 1735 | 1718 | 0.102 | 0.234 | 0.077 | -0.002 | -0.073 |
|---|
| THBS2 |   | 3 | 557 | 1903 | 1868 | 0.094 | 0.221 | 0.060 | 0.241 | 0.154 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| USP8 |   | 1 | 1195 | 1903 | 1915 | -0.137 | 0.046 | 0.083 | 0.015 | 0.139 |
|---|
| GNG4 |   | 5 | 360 | 1077 | 1057 | 0.095 | -0.139 | -0.169 | 0.129 | 0.106 |
|---|
| SUPT5H |   | 6 | 301 | 1323 | 1281 | -0.085 | 0.007 | -0.298 | 0.002 | -0.068 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| FANCA |   | 9 | 196 | 1 | 44 | 0.120 | -0.188 | 0.106 | -0.127 | -0.053 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| POLR2A |   | 6 | 301 | 1547 | 1510 | -0.107 | 0.247 | 0.091 | 0.040 | 0.141 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| RAI14 |   | 2 | 743 | 1903 | 1878 | 0.063 | 0.188 | 0.114 | undef | 0.225 |
|---|
| POLR2E |   | 2 | 743 | 1396 | 1391 | 0.127 | 0.039 | 0.166 | 0.084 | -0.018 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
GO Enrichment output for subnetwork 6267 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sarcomere organization | GO:0045214 |  | 8.413E-16 | 2.055E-12 |
|---|
| actomyosin structure organization | GO:0031032 |  | 2.667E-15 | 3.257E-12 |
|---|
| cellular component assembly involved in morphogenesis | GO:0010927 |  | 3.603E-15 | 2.934E-12 |
|---|
| myofibril assembly | GO:0030239 |  | 2.337E-14 | 1.427E-11 |
|---|
| muscle cell development | GO:0055001 |  | 4.543E-14 | 2.22E-11 |
|---|
| response to calcium ion | GO:0051592 |  | 5.971E-14 | 2.431E-11 |
|---|
| response to metal ion | GO:0010038 |  | 5.563E-13 | 1.941E-10 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 8.707E-13 | 2.659E-10 |
|---|
| response to inorganic substance | GO:0010035 |  | 1.214E-12 | 3.295E-10 |
|---|
| muscle cell differentiation | GO:0042692 |  | 3.416E-10 | 8.346E-08 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.168E-09 | 7.036E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sarcomere organization | GO:0045214 |  | 1.969E-15 | 4.736E-12 |
|---|
| cellular component assembly involved in morphogenesis | GO:0010927 |  | 6.807E-15 | 8.189E-12 |
|---|
| actomyosin structure organization | GO:0031032 |  | 6.807E-15 | 5.459E-12 |
|---|
| myofibril assembly | GO:0030239 |  | 5.45E-14 | 3.278E-11 |
|---|
| response to calcium ion | GO:0051592 |  | 9.647E-14 | 4.642E-11 |
|---|
| muscle cell development | GO:0055001 |  | 1.058E-13 | 4.244E-11 |
|---|
| response to metal ion | GO:0010038 |  | 8.785E-13 | 3.019E-10 |
|---|
| response to inorganic substance | GO:0010035 |  | 1.993E-12 | 5.994E-10 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 2.019E-12 | 5.398E-10 |
|---|
| muscle cell differentiation | GO:0042692 |  | 6.963E-10 | 1.675E-07 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 4.651E-09 | 1.017E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sarcomere organization | GO:0045214 |  | 3.948E-15 | 9.081E-12 |
|---|
| cellular component assembly involved in morphogenesis | GO:0010927 |  | 1.206E-14 | 1.387E-11 |
|---|
| actomyosin structure organization | GO:0031032 |  | 2.487E-14 | 1.906E-11 |
|---|
| myofibril assembly | GO:0030239 |  | 1.907E-13 | 1.097E-10 |
|---|
| response to calcium ion | GO:0051592 |  | 2.181E-13 | 1.003E-10 |
|---|
| muscle cell development | GO:0055001 |  | 4.025E-13 | 1.543E-10 |
|---|
| response to metal ion | GO:0010038 |  | 3.145E-12 | 1.033E-09 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 6.199E-12 | 1.782E-09 |
|---|
| response to inorganic substance | GO:0010035 |  | 6.904E-12 | 1.764E-09 |
|---|
| muscle cell differentiation | GO:0042692 |  | 2.312E-09 | 5.318E-07 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.363E-08 | 2.85E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.319E-07 | 4.273E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.875E-07 | 6.336E-04 |
|---|
| response to hormone stimulus | GO:0009725 |  | 9.005E-07 | 5.532E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.295E-06 | 5.965E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.144E-06 | 7.903E-04 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 2.3E-06 | 7.066E-04 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 4.341E-06 | 1.143E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.521E-06 | 1.042E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 4.521E-06 | 9.258E-04 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.521E-06 | 8.333E-04 |
|---|
| cell junction assembly | GO:0034329 |  | 7.48E-06 | 1.253E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sarcomere organization | GO:0045214 |  | 3.948E-15 | 9.081E-12 |
|---|
| cellular component assembly involved in morphogenesis | GO:0010927 |  | 1.206E-14 | 1.387E-11 |
|---|
| actomyosin structure organization | GO:0031032 |  | 2.487E-14 | 1.906E-11 |
|---|
| myofibril assembly | GO:0030239 |  | 1.907E-13 | 1.097E-10 |
|---|
| response to calcium ion | GO:0051592 |  | 2.181E-13 | 1.003E-10 |
|---|
| muscle cell development | GO:0055001 |  | 4.025E-13 | 1.543E-10 |
|---|
| response to metal ion | GO:0010038 |  | 3.145E-12 | 1.033E-09 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 6.199E-12 | 1.782E-09 |
|---|
| response to inorganic substance | GO:0010035 |  | 6.904E-12 | 1.764E-09 |
|---|
| muscle cell differentiation | GO:0042692 |  | 2.312E-09 | 5.318E-07 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.363E-08 | 2.85E-06 |
|---|



