Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6209
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7599 | 3.154e-03 | 4.970e-04 | 7.544e-01 | 1.182e-06 |
|---|
| Loi | 0.2279 | 8.409e-02 | 1.239e-02 | 4.374e-01 | 4.557e-04 |
|---|
| Schmidt | 0.6720 | 0.000e+00 | 0.000e+00 | 4.647e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.506e-03 | 0.000e+00 |
|---|
| Wang | 0.2593 | 3.007e-03 | 4.674e-02 | 4.097e-01 | 5.758e-05 |
|---|
Expression data for subnetwork 6209 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
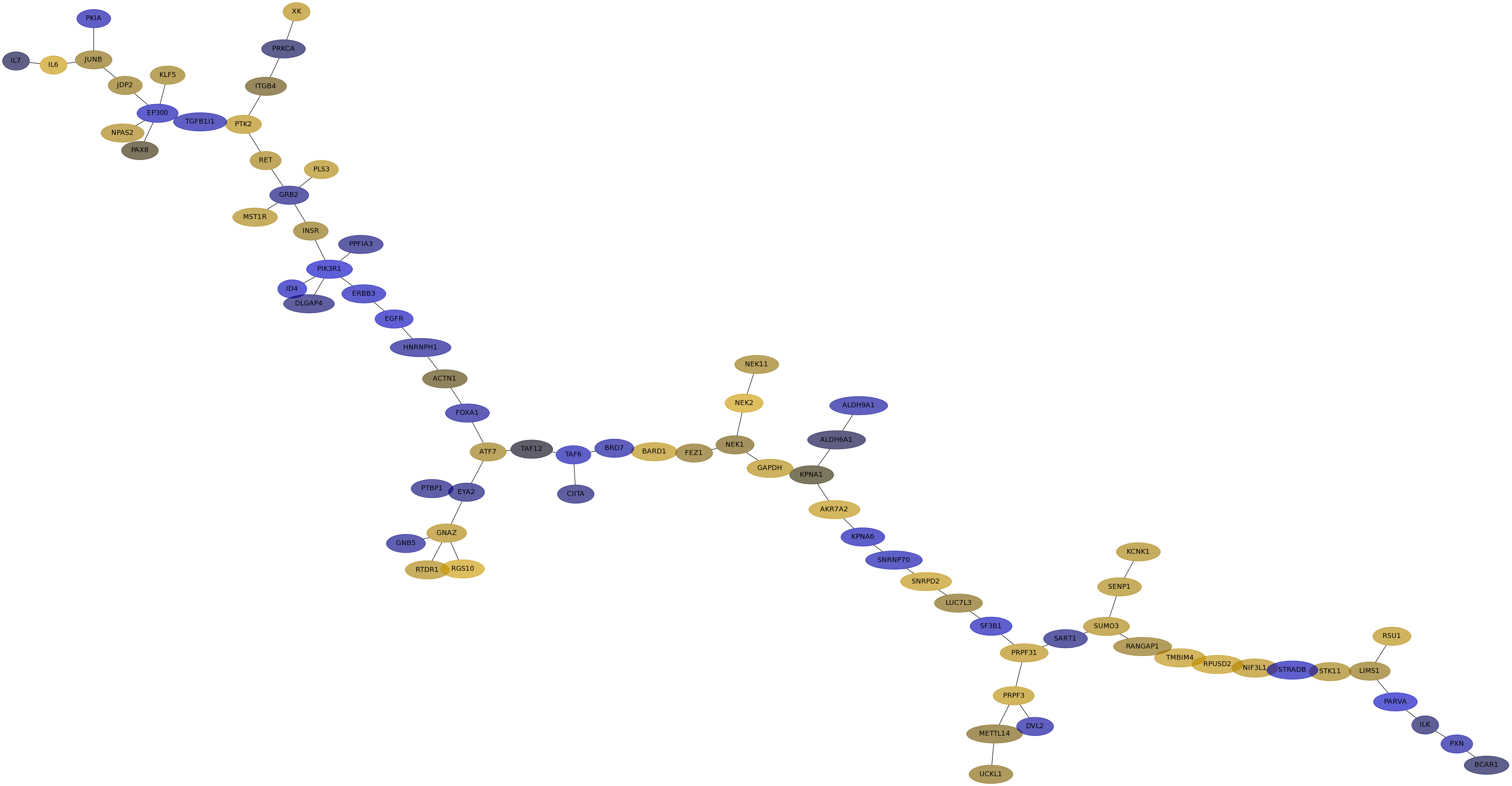
Score for each gene in subnetwork 6209 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SART1 |   | 1 | 1195 | 2053 | 2058 | -0.035 | -0.166 | -0.085 | 0.018 | 0.028 |
|---|
| NPAS2 |   | 3 | 557 | 366 | 389 | 0.116 | 0.253 | -0.157 | -0.062 | -0.093 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| RET |   | 7 | 256 | 366 | 376 | 0.098 | 0.063 | -0.179 | undef | -0.159 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| RPUSD2 |   | 1 | 1195 | 2053 | 2058 | 0.209 | -0.015 | 0.199 | undef | 0.169 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| PLS3 |   | 1 | 1195 | 2053 | 2058 | 0.144 | 0.175 | 0.247 | 0.120 | 0.256 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| METTL14 |   | 2 | 743 | 1928 | 1911 | 0.035 | -0.039 | undef | undef | undef |
|---|
| SENP1 |   | 2 | 743 | 1364 | 1363 | 0.126 | -0.034 | undef | -0.075 | undef |
|---|
| DVL2 |   | 1 | 1195 | 2053 | 2058 | -0.097 | -0.001 | -0.198 | undef | 0.124 |
|---|
| NIF3L1 |   | 1 | 1195 | 2053 | 2058 | 0.153 | -0.130 | -0.113 | undef | 0.013 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| LIMS1 |   | 1 | 1195 | 2053 | 2058 | 0.063 | 0.090 | -0.032 | 0.059 | 0.277 |
|---|
| PTBP1 |   | 3 | 557 | 1165 | 1151 | -0.038 | -0.090 | -0.113 | 0.054 | -0.110 |
|---|
| UCKL1 |   | 1 | 1195 | 2053 | 2058 | 0.050 | -0.023 | 0.164 | 0.076 | -0.069 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| SUMO3 |   | 3 | 557 | 412 | 430 | 0.126 | 0.081 | 0.087 | 0.138 | -0.073 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| PPFIA3 |   | 2 | 743 | 2053 | 2031 | -0.037 | -0.042 | -0.144 | undef | 0.014 |
|---|
| PRPF3 |   | 2 | 743 | 1928 | 1911 | 0.186 | -0.006 | 0.053 | 0.048 | -0.058 |
|---|
| PARVA |   | 1 | 1195 | 2053 | 2058 | -0.214 | 0.082 | 0.114 | 0.226 | 0.263 |
|---|
| RSU1 |   | 1 | 1195 | 2053 | 2058 | 0.168 | -0.198 | 0.051 | 0.063 | 0.085 |
|---|
| MST1R |   | 5 | 360 | 236 | 242 | 0.124 | 0.083 | -0.212 | -0.001 | 0.026 |
|---|
| KCNK1 |   | 1 | 1195 | 2053 | 2058 | 0.119 | 0.214 | -0.329 | 0.287 | 0.037 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| RGS10 |   | 2 | 743 | 842 | 854 | 0.295 | -0.214 | -0.002 | undef | -0.095 |
|---|
| STK11 |   | 2 | 743 | 2053 | 2031 | 0.102 | -0.125 | -0.080 | -0.022 | -0.079 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| RTDR1 |   | 5 | 360 | 1364 | 1328 | 0.135 | 0.098 | 0.222 | 0.125 | undef |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| ALDH6A1 |   | 1 | 1195 | 2053 | 2058 | -0.010 | 0.124 | 0.000 | undef | 0.094 |
|---|
| TMBIM4 |   | 1 | 1195 | 2053 | 2058 | 0.197 | 0.017 | -0.160 | 0.092 | 0.169 |
|---|
| PRPF31 |   | 1 | 1195 | 2053 | 2058 | 0.167 | 0.076 | 0.119 | -0.120 | -0.089 |
|---|
| JDP2 |   | 4 | 440 | 2053 | 2008 | 0.062 | 0.045 | undef | undef | undef |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| ERBB3 |   | 3 | 557 | 1922 | 1888 | -0.171 | -0.007 | -0.141 | -0.019 | -0.098 |
|---|
| AKR7A2 |   | 1 | 1195 | 2053 | 2058 | 0.211 | 0.032 | -0.017 | 0.025 | 0.021 |
|---|
| KPNA6 |   | 1 | 1195 | 2053 | 2058 | -0.144 | 0.128 | -0.005 | -0.246 | -0.108 |
|---|
| ALDH9A1 |   | 1 | 1195 | 2053 | 2058 | -0.089 | 0.265 | -0.199 | 0.122 | -0.020 |
|---|
| IL7 |   | 1 | 1195 | 2053 | 2058 | -0.010 | 0.110 | -0.156 | -0.042 | 0.077 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| STRADB |   | 1 | 1195 | 2053 | 2058 | -0.157 | -0.105 | undef | 0.116 | undef |
|---|
| RANGAP1 |   | 1 | 1195 | 2053 | 2058 | 0.060 | 0.019 | 0.063 | 0.089 | 0.080 |
|---|
| PTK2 |   | 4 | 440 | 1437 | 1414 | 0.166 | -0.056 | 0.160 | -0.032 | -0.020 |
|---|
| ILK |   | 4 | 440 | 1413 | 1390 | -0.020 | 0.195 | 0.076 | 0.130 | 0.086 |
|---|
| BCAR1 |   | 6 | 301 | 1 | 52 | -0.012 | 0.229 | undef | 0.223 | undef |
|---|
| CIITA |   | 1 | 1195 | 2053 | 2058 | -0.029 | -0.140 | 0.017 | -0.147 | -0.108 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| PAX8 |   | 1 | 1195 | 2053 | 2058 | 0.008 | -0.083 | 0.032 | -0.227 | -0.002 |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| NEK11 |   | 1 | 1195 | 2053 | 2058 | 0.081 | 0.152 | -0.197 | undef | 0.040 |
|---|
| TAF12 |   | 1 | 1195 | 2053 | 2058 | -0.003 | -0.098 | -0.076 | 0.115 | -0.077 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| SNRNP70 |   | 5 | 360 | 296 | 310 | -0.129 | 0.130 | -0.059 | -0.099 | 0.036 |
|---|
| SNRPD2 |   | 1 | 1195 | 2053 | 2058 | 0.210 | -0.181 | 0.085 | undef | -0.045 |
|---|
| TAF6 |   | 2 | 743 | 457 | 481 | -0.139 | -0.013 | -0.015 | 0.203 | 0.073 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
GO Enrichment output for subnetwork 6209 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.346E-07 | 1.306E-03 |
|---|
| negative regulation of transport | GO:0051051 |  | 8.352E-07 | 1.02E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.15E-06 | 2.565E-03 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 6.927E-06 | 4.23E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 7.296E-06 | 3.565E-03 |
|---|
| cell aging | GO:0007569 |  | 8.426E-06 | 3.431E-03 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 8.426E-06 | 2.941E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 9.215E-06 | 2.814E-03 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.015E-05 | 2.756E-03 |
|---|
| regulation of behavior | GO:0050795 |  | 1.437E-05 | 3.51E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.736E-05 | 3.855E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 6.05E-07 | 1.456E-03 |
|---|
| negative regulation of transport | GO:0051051 |  | 1.188E-06 | 1.43E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.786E-06 | 3.037E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.899E-06 | 5.353E-03 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 8.983E-06 | 4.323E-03 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 8.983E-06 | 3.602E-03 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.092E-05 | 3.755E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.136E-05 | 3.417E-03 |
|---|
| regulation of behavior | GO:0050795 |  | 1.571E-05 | 4.2E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.116E-05 | 5.09E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.556E-05 | 5.591E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of transport | GO:0051051 |  | 2.409E-06 | 5.54E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.909E-05 | 0.02195444 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.984E-05 | 0.01520985 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.984E-05 | 0.01140739 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 2.435E-05 | 0.01120069 |
|---|
| regulation of behavior | GO:0050795 |  | 2.957E-05 | 0.01133477 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.036E-05 | 9.975E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.24E-05 | 0.01219072 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 4.526E-05 | 0.01156701 |
|---|
| positive regulation of T cell differentiation | GO:0045582 |  | 7.963E-05 | 0.01831389 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 8.783E-05 | 0.01836453 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.725E-06 | 6.866E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.399E-06 | 5.897E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 8.212E-06 | 5.045E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 9.607E-06 | 4.427E-03 |
|---|
| positive regulation of T cell differentiation | GO:0045582 |  | 1.034E-05 | 3.813E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.58E-05 | 4.852E-03 |
|---|
| positive regulation of lymphocyte differentiation | GO:0045621 |  | 1.92E-05 | 5.054E-03 |
|---|
| negative regulation of transport | GO:0051051 |  | 3.32E-05 | 7.65E-03 |
|---|
| regulation of T cell differentiation | GO:0045580 |  | 3.837E-05 | 7.857E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.837E-05 | 7.071E-03 |
|---|
| negative regulation of nucleocytoplasmic transport | GO:0046823 |  | 4.708E-05 | 7.889E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of transport | GO:0051051 |  | 2.409E-06 | 5.54E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.909E-05 | 0.02195444 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.984E-05 | 0.01520985 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.984E-05 | 0.01140739 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 2.435E-05 | 0.01120069 |
|---|
| regulation of behavior | GO:0050795 |  | 2.957E-05 | 0.01133477 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.036E-05 | 9.975E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.24E-05 | 0.01219072 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 4.526E-05 | 0.01156701 |
|---|
| positive regulation of T cell differentiation | GO:0045582 |  | 7.963E-05 | 0.01831389 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 8.783E-05 | 0.01836453 |
|---|



