Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6151
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7608 | 3.102e-03 | 4.870e-04 | 7.518e-01 | 1.136e-06 |
|---|
| Loi | 0.2262 | 8.678e-02 | 1.314e-02 | 4.438e-01 | 5.060e-04 |
|---|
| Schmidt | 0.6730 | 0.000e+00 | 0.000e+00 | 4.535e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7902 | 0.000e+00 | 0.000e+00 | 3.544e-03 | 0.000e+00 |
|---|
| Wang | 0.2592 | 3.012e-03 | 4.678e-02 | 4.099e-01 | 5.775e-05 |
|---|
Expression data for subnetwork 6151 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
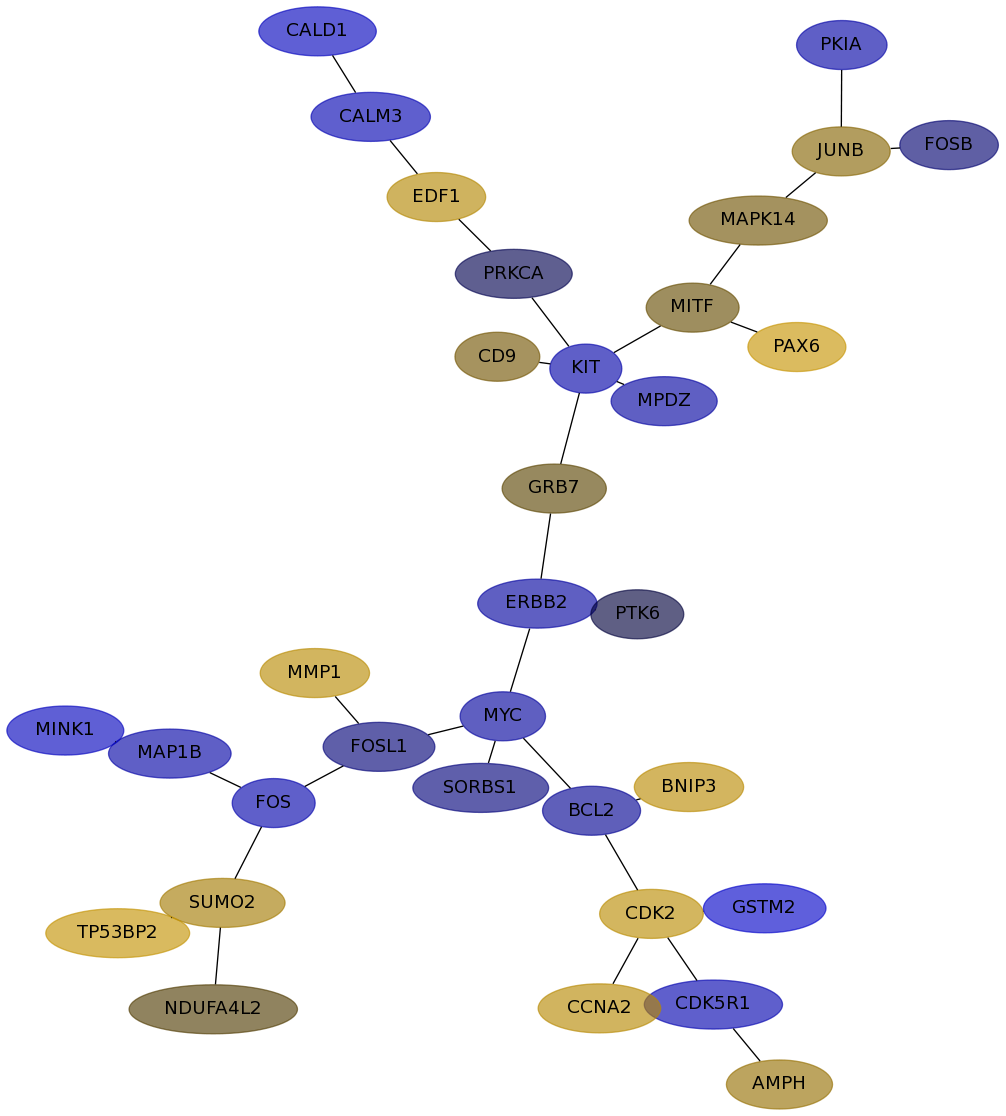
Score for each gene in subnetwork 6151 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PTK6 |   | 3 | 557 | 2141 | 2102 | -0.010 | 0.204 | -0.343 | 0.069 | 0.213 |
|---|
| MINK1 |   | 2 | 743 | 2141 | 2117 | -0.210 | -0.166 | -0.092 | 0.103 | -0.117 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| CALM3 |   | 1 | 1195 | 2141 | 2147 | -0.163 | 0.107 | 0.007 | -0.204 | -0.003 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| MAPK14 |   | 4 | 440 | 1 | 59 | 0.033 | -0.065 | 0.299 | 0.015 | 0.119 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| AMPH |   | 7 | 256 | 83 | 102 | 0.082 | 0.000 | -0.104 | 0.024 | 0.241 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| CALD1 |   | 1 | 1195 | 2141 | 2147 | -0.209 | 0.242 | 0.032 | undef | 0.229 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| FOSB |   | 5 | 360 | 1534 | 1501 | -0.033 | 0.046 | 0.129 | -0.104 | 0.095 |
|---|
| MITF |   | 5 | 360 | 696 | 680 | 0.027 | 0.032 | 0.065 | 0.145 | 0.183 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| MPDZ |   | 3 | 557 | 1846 | 1817 | -0.110 | 0.062 | 0.231 | 0.291 | 0.167 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| NDUFA4L2 |   | 2 | 743 | 1928 | 1911 | 0.015 | -0.030 | 0.065 | 0.139 | 0.174 |
|---|
| GSTM2 |   | 1 | 1195 | 2141 | 2147 | -0.271 | -0.138 | 0.010 | 0.020 | -0.088 |
|---|
| MMP1 |   | 11 | 148 | 366 | 360 | 0.192 | 0.014 | 0.036 | 0.044 | 0.260 |
|---|
| EDF1 |   | 1 | 1195 | 2141 | 2147 | 0.168 | 0.250 | 0.058 | undef | -0.098 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| FOSL1 |   | 11 | 148 | 366 | 360 | -0.040 | 0.164 | 0.010 | 0.128 | 0.005 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
GO Enrichment output for subnetwork 6151 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pigmentation | GO:0043473 |  | 9.361E-10 | 2.287E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.094E-07 | 1.337E-04 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 1.094E-07 | 8.911E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.818E-07 | 2.332E-04 |
|---|
| cell junction assembly | GO:0034329 |  | 7.146E-07 | 3.492E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.133E-07 | 3.718E-04 |
|---|
| T cell selection | GO:0045058 |  | 9.133E-07 | 3.187E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.303E-06 | 3.978E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 1.303E-06 | 3.536E-04 |
|---|
| cell junction organization | GO:0034330 |  | 1.485E-06 | 3.627E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 1.788E-06 | 3.971E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pigmentation | GO:0043473 |  | 1.977E-09 | 4.757E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.601E-07 | 1.926E-04 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 1.601E-07 | 1.284E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 5.583E-07 | 3.358E-04 |
|---|
| cell junction assembly | GO:0034329 |  | 1.029E-06 | 4.951E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.335E-06 | 5.353E-04 |
|---|
| T cell selection | GO:0045058 |  | 1.335E-06 | 4.588E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.903E-06 | 5.725E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 1.903E-06 | 5.089E-04 |
|---|
| cell junction organization | GO:0034330 |  | 2.186E-06 | 5.259E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 2.612E-06 | 5.714E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pigmentation | GO:0043473 |  | 3.549E-09 | 8.163E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 4.022E-07 | 4.626E-04 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 4.022E-07 | 3.084E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.401E-06 | 8.055E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.236E-06 | 1.028E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 2.58E-06 | 9.889E-04 |
|---|
| T cell selection | GO:0045058 |  | 3.345E-06 | 1.099E-03 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.345E-06 | 9.617E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 4.767E-06 | 1.218E-03 |
|---|
| cell junction organization | GO:0034330 |  | 5.089E-06 | 1.17E-03 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.538E-06 | 1.367E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell junction assembly | GO:0034329 |  | 2.63E-07 | 4.848E-04 |
|---|
| pigmentation | GO:0043473 |  | 2.63E-07 | 2.424E-04 |
|---|
| regulation of membrane potential | GO:0042391 |  | 4.132E-07 | 2.538E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 7.41E-07 | 3.414E-04 |
|---|
| cell junction organization | GO:0034330 |  | 7.991E-07 | 2.946E-04 |
|---|
| T cell selection | GO:0045058 |  | 2.065E-06 | 6.344E-04 |
|---|
| pigmentation during development | GO:0048066 |  | 3.091E-06 | 8.138E-04 |
|---|
| T cell differentiation in the thymus | GO:0033077 |  | 3.091E-06 | 7.121E-04 |
|---|
| defense response to virus | GO:0051607 |  | 3.091E-06 | 6.33E-04 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 3.212E-06 | 5.92E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 4.406E-06 | 7.381E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pigmentation | GO:0043473 |  | 3.549E-09 | 8.163E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 4.022E-07 | 4.626E-04 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 4.022E-07 | 3.084E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.401E-06 | 8.055E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.236E-06 | 1.028E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 2.58E-06 | 9.889E-04 |
|---|
| T cell selection | GO:0045058 |  | 3.345E-06 | 1.099E-03 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.345E-06 | 9.617E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 4.767E-06 | 1.218E-03 |
|---|
| cell junction organization | GO:0034330 |  | 5.089E-06 | 1.17E-03 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.538E-06 | 1.367E-03 |
|---|



