Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6134
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7612 | 3.078e-03 | 4.820e-04 | 7.505e-01 | 1.113e-06 |
|---|
| Loi | 0.2258 | 8.740e-02 | 1.332e-02 | 4.452e-01 | 5.182e-04 |
|---|
| Schmidt | 0.6731 | 0.000e+00 | 0.000e+00 | 4.526e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7902 | 0.000e+00 | 0.000e+00 | 3.547e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.031e-03 | 4.695e-02 | 4.106e-01 | 5.842e-05 |
|---|
Expression data for subnetwork 6134 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
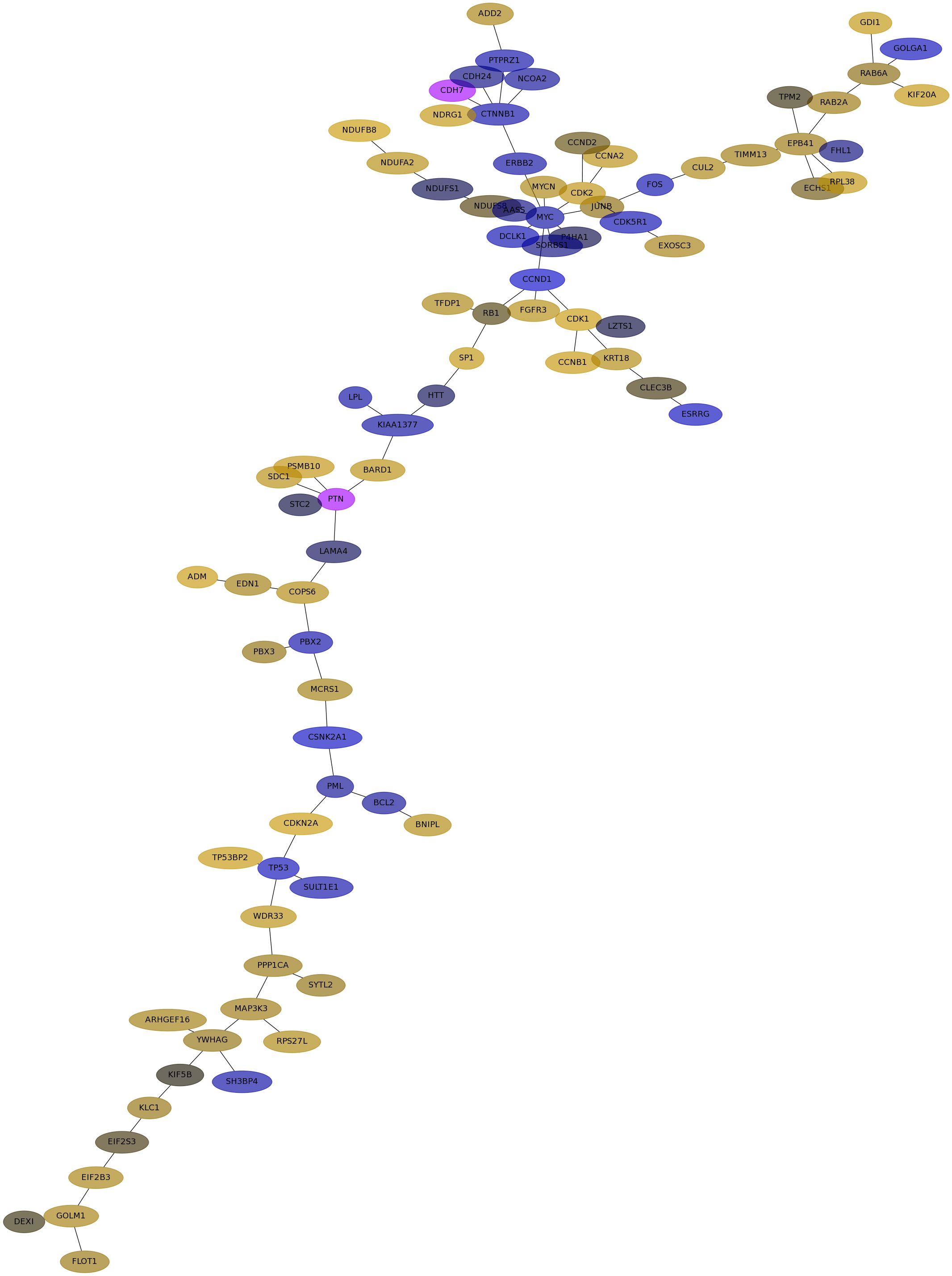
Score for each gene in subnetwork 6134 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| AASS |   | 2 | 743 | 2147 | 2126 | -0.047 | 0.229 | 0.021 | 0.187 | 0.002 |
|---|
| NDRG1 |   | 6 | 301 | 658 | 646 | 0.222 | 0.143 | -0.040 | -0.018 | 0.242 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| CSNK2A1 |   | 3 | 557 | 1510 | 1486 | -0.215 | 0.029 | undef | 0.122 | undef |
|---|
| RAB2A |   | 1 | 1195 | 2147 | 2151 | 0.083 | 0.049 | -0.129 | 0.211 | 0.191 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CCNB1 |   | 4 | 440 | 1116 | 1101 | 0.241 | -0.056 | 0.001 | undef | -0.043 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| SDC1 |   | 2 | 743 | 1954 | 1933 | 0.172 | 0.241 | -0.088 | 0.179 | 0.059 |
|---|
| SYTL2 |   | 1 | 1195 | 2147 | 2151 | 0.063 | 0.186 | undef | undef | undef |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| GOLGA1 |   | 1 | 1195 | 2147 | 2151 | -0.179 | 0.089 | 0.081 | 0.051 | -0.064 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| NDUFB8 |   | 5 | 360 | 696 | 680 | 0.286 | -0.012 | 0.340 | -0.171 | 0.205 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| PPP1CA |   | 2 | 743 | 1621 | 1611 | 0.076 | -0.061 | 0.020 | 0.196 | -0.168 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| MAP3K3 |   | 4 | 440 | 1364 | 1330 | 0.085 | 0.046 | 0.028 | -0.035 | 0.094 |
|---|
| RPS27L |   | 2 | 743 | 1364 | 1363 | 0.131 | 0.039 | -0.033 | -0.098 | 0.026 |
|---|
| RPL38 |   | 2 | 743 | 141 | 197 | 0.222 | 0.151 | 0.241 | -0.203 | -0.171 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| BNIPL |   | 2 | 743 | 1735 | 1721 | 0.144 | 0.072 | undef | undef | undef |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| EPB41 |   | 2 | 743 | 318 | 346 | 0.082 | -0.247 | 0.000 | -0.179 | -0.042 |
|---|
| TIMM13 |   | 1 | 1195 | 2147 | 2151 | 0.091 | -0.078 | 0.164 | undef | 0.089 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| P4HA1 |   | 2 | 743 | 957 | 958 | -0.011 | 0.089 | -0.020 | undef | 0.169 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| EIF2B3 |   | 1 | 1195 | 2147 | 2151 | 0.115 | -0.039 | -0.013 | undef | -0.110 |
|---|
| LZTS1 |   | 3 | 557 | 2147 | 2108 | -0.008 | -0.089 | 0.043 | 0.147 | 0.063 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| NDUFS1 |   | 1 | 1195 | 2147 | 2151 | -0.013 | -0.045 | -0.123 | undef | 0.061 |
|---|
| EXOSC3 |   | 3 | 557 | 552 | 558 | 0.107 | 0.121 | undef | -0.074 | undef |
|---|
| SULT1E1 |   | 1 | 1195 | 2147 | 2151 | -0.120 | -0.008 | -0.110 | -0.040 | 0.245 |
|---|
| FLOT1 |   | 2 | 743 | 1102 | 1102 | 0.076 | 0.004 | -0.095 | 0.188 | -0.095 |
|---|
| PBX3 |   | 2 | 743 | 983 | 988 | 0.065 | 0.008 | 0.034 | 0.103 | 0.021 |
|---|
| KLC1 |   | 4 | 440 | 1055 | 1035 | 0.071 | 0.189 | 0.204 | 0.139 | 0.132 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| RAB6A |   | 2 | 743 | 842 | 854 | 0.054 | -0.037 | -0.038 | 0.208 | -0.164 |
|---|
| GDI1 |   | 3 | 557 | 842 | 834 | 0.219 | 0.094 | 0.049 | 0.181 | 0.049 |
|---|
| KIF5B |   | 2 | 743 | 2111 | 2085 | 0.003 | 0.010 | -0.104 | 0.082 | -0.149 |
|---|
| GOLM1 |   | 8 | 222 | 1534 | 1489 | 0.109 | 0.161 | -0.217 | 0.170 | -0.060 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| CDH7 |   | 1 | 1195 | 2147 | 2151 | undef | -0.025 | -0.088 | 0.162 | 0.156 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| NDUFA2 |   | 2 | 743 | 696 | 717 | 0.159 | -0.014 | 0.084 | -0.037 | -0.022 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| STC2 |   | 5 | 360 | 236 | 242 | -0.009 | 0.056 | 0.181 | -0.185 | 0.016 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| NDUFS8 |   | 3 | 557 | 696 | 695 | 0.013 | -0.088 | -0.150 | 0.176 | 0.030 |
|---|
| MYCN |   | 7 | 256 | 764 | 756 | 0.125 | 0.057 | -0.003 | -0.081 | -0.084 |
|---|
| CCND2 |   | 5 | 360 | 1492 | 1453 | 0.022 | -0.011 | -0.072 | -0.153 | -0.029 |
|---|
| NCOA2 |   | 3 | 557 | 1165 | 1151 | -0.078 | 0.080 | -0.159 | 0.104 | 0.057 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MCRS1 |   | 1 | 1195 | 2147 | 2151 | 0.098 | -0.047 | -0.039 | 0.051 | -0.169 |
|---|
| TPM2 |   | 4 | 440 | 1034 | 1015 | 0.007 | 0.269 | 0.212 | 0.051 | 0.203 |
|---|
| KIF20A |   | 1 | 1195 | 2147 | 2151 | 0.233 | -0.083 | 0.073 | -0.035 | -0.005 |
|---|
| ESRRG |   | 6 | 301 | 614 | 602 | -0.205 | 0.197 | -0.007 | 0.155 | -0.085 |
|---|
| ECHS1 |   | 2 | 743 | 1190 | 1189 | 0.028 | 0.053 | 0.156 | 0.205 | -0.042 |
|---|
| EDN1 |   | 6 | 301 | 682 | 669 | 0.100 | 0.126 | 0.249 | -0.086 | 0.121 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| PBX2 |   | 5 | 360 | 1 | 56 | -0.122 | -0.073 | 0.264 | -0.095 | 0.092 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| DEXI |   | 1 | 1195 | 2147 | 2151 | 0.007 | -0.053 | 0.145 | 0.079 | 0.209 |
|---|
| CDH24 |   | 1 | 1195 | 2147 | 2151 | -0.048 | 0.141 | undef | undef | undef |
|---|
| PSMB10 |   | 1 | 1195 | 2147 | 2151 | 0.213 | -0.213 | -0.123 | -0.276 | -0.046 |
|---|
| EIF2S3 |   | 1 | 1195 | 2147 | 2151 | 0.009 | -0.092 | 0.011 | undef | -0.104 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| TFDP1 |   | 6 | 301 | 806 | 783 | 0.123 | -0.178 | 0.129 | undef | -0.065 |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| SH3BP4 |   | 4 | 440 | 957 | 942 | -0.105 | 0.108 | -0.100 | 0.246 | -0.003 |
|---|
| PML |   | 6 | 301 | 478 | 473 | -0.068 | -0.010 | -0.007 | -0.153 | -0.070 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| WDR33 |   | 2 | 743 | 1364 | 1363 | 0.174 | -0.117 | 0.001 | -0.067 | 0.083 |
|---|
| KRT18 |   | 8 | 222 | 590 | 569 | 0.148 | 0.100 | -0.083 | 0.024 | -0.053 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CLEC3B |   | 2 | 743 | 928 | 929 | 0.009 | -0.021 | 0.139 | undef | 0.033 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6134 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.797E-08 | 4.39E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.885E-08 | 2.302E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.108E-08 | 1.716E-05 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 6.309E-08 | 3.853E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.695E-07 | 8.28E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 3.727E-07 | 1.518E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 4.554E-07 | 1.589E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.74E-06 | 5.314E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 1.83E-06 | 4.968E-04 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 2.361E-06 | 5.767E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.79E-06 | 6.196E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.928E-08 | 7.044E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.189E-08 | 3.836E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.738E-08 | 2.998E-05 |
|---|
| interphase | GO:0051325 |  | 3.849E-08 | 2.315E-05 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 8.856E-08 | 4.261E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.573E-07 | 1.032E-04 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 5.224E-07 | 1.796E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 6.553E-07 | 1.971E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.435E-06 | 6.509E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 2.944E-06 | 7.083E-04 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 3.301E-06 | 7.221E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.191E-08 | 1.194E-04 |
|---|
| interphase | GO:0051325 |  | 6.356E-08 | 7.31E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.401E-08 | 5.674E-05 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 1.786E-07 | 1.027E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.406E-07 | 1.567E-04 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 1.051E-06 | 4.027E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.089E-06 | 3.578E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.227E-06 | 3.529E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 3.019E-06 | 7.714E-04 |
|---|
| ear development | GO:0043583 |  | 4.201E-06 | 9.663E-04 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 4.875E-06 | 1.019E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 9.247E-09 | 1.704E-05 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 9.448E-08 | 8.706E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 4.349E-07 | 2.672E-04 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 5.773E-07 | 2.66E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.961E-07 | 3.672E-04 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 9.961E-07 | 3.06E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.62E-06 | 4.264E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.624E-06 | 3.742E-04 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 2.008E-06 | 4.112E-04 |
|---|
| regulation of cell size | GO:0008361 |  | 2.402E-06 | 4.426E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.109E-06 | 5.208E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.191E-08 | 1.194E-04 |
|---|
| interphase | GO:0051325 |  | 6.356E-08 | 7.31E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.401E-08 | 5.674E-05 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 1.786E-07 | 1.027E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.406E-07 | 1.567E-04 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 1.051E-06 | 4.027E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.089E-06 | 3.578E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.227E-06 | 3.529E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 3.019E-06 | 7.714E-04 |
|---|
| ear development | GO:0043583 |  | 4.201E-06 | 9.663E-04 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 4.875E-06 | 1.019E-03 |
|---|



