Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6119
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7615 | 3.061e-03 | 4.790e-04 | 7.496e-01 | 1.099e-06 |
|---|
| Loi | 0.2254 | 8.809e-02 | 1.351e-02 | 4.468e-01 | 5.318e-04 |
|---|
| Schmidt | 0.6732 | 0.000e+00 | 0.000e+00 | 4.516e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7901 | 0.000e+00 | 0.000e+00 | 3.553e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.031e-03 | 4.695e-02 | 4.106e-01 | 5.843e-05 |
|---|
Expression data for subnetwork 6119 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
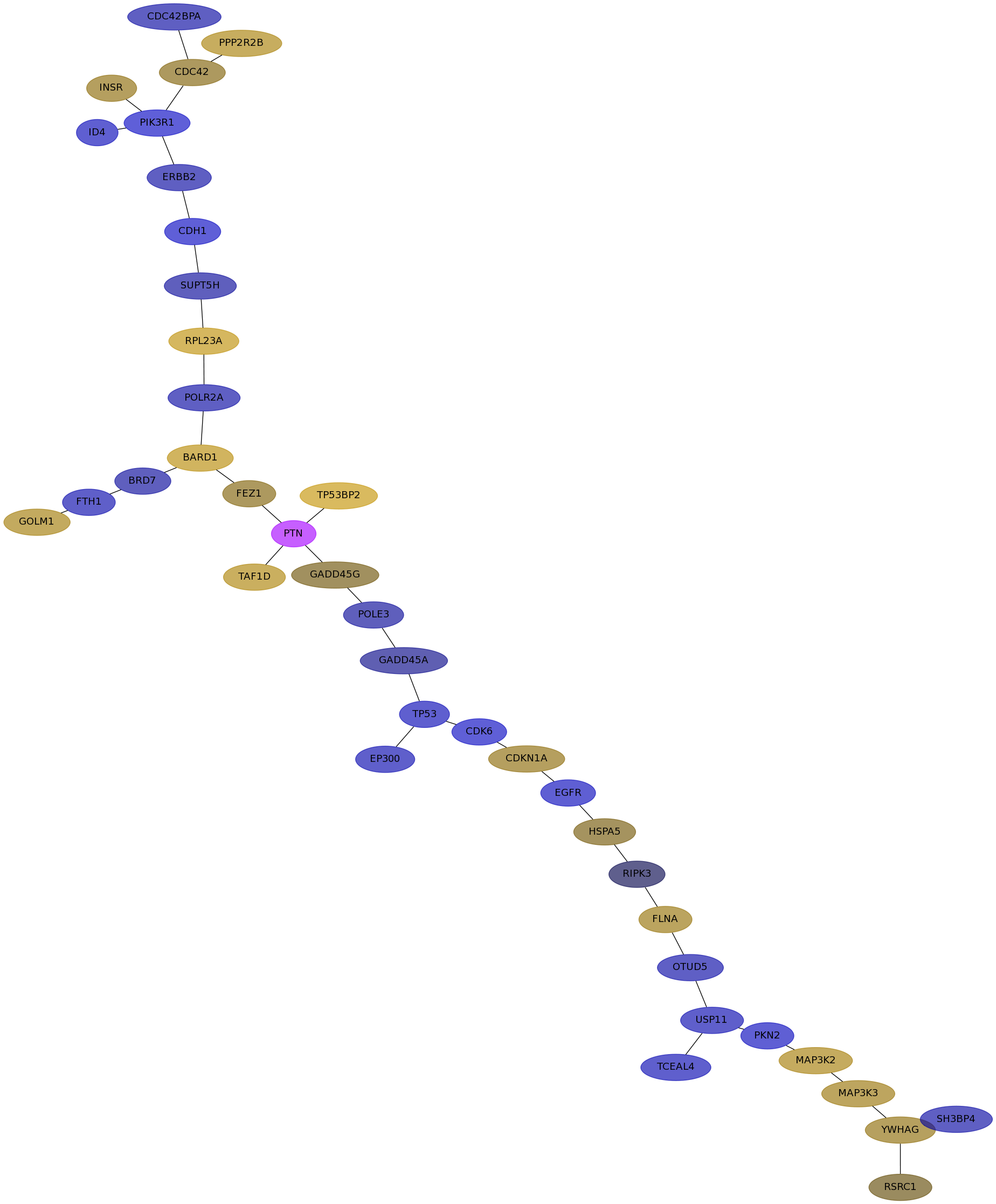
Score for each gene in subnetwork 6119 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| OTUD5 |   | 1 | 1195 | 2235 | 2236 | -0.113 | -0.152 | undef | 0.094 | undef |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| FTH1 |   | 1 | 1195 | 2235 | 2236 | -0.135 | 0.073 | -0.200 | -0.014 | -0.193 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| RPL23A |   | 5 | 360 | 842 | 828 | 0.208 | -0.007 | 0.024 | -0.242 | 0.055 |
|---|
| GOLM1 |   | 8 | 222 | 1534 | 1489 | 0.109 | 0.161 | -0.217 | 0.170 | -0.060 |
|---|
| PKN2 |   | 1 | 1195 | 2235 | 2236 | -0.198 | 0.158 | 0.060 | -0.092 | 0.068 |
|---|
| MAP3K2 |   | 2 | 743 | 2215 | 2193 | 0.115 | 0.087 | -0.066 | -0.070 | -0.117 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| MAP3K3 |   | 4 | 440 | 1364 | 1330 | 0.085 | 0.046 | 0.028 | -0.035 | 0.094 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| HSPA5 |   | 2 | 743 | 2180 | 2165 | 0.035 | 0.033 | -0.006 | undef | -0.203 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDC42BPA |   | 6 | 301 | 1268 | 1236 | -0.102 | 0.098 | 0.059 | 0.368 | 0.260 |
|---|
| SUPT5H |   | 6 | 301 | 1323 | 1281 | -0.085 | 0.007 | -0.298 | 0.002 | -0.068 |
|---|
| POLR2A |   | 6 | 301 | 1547 | 1510 | -0.107 | 0.247 | 0.091 | 0.040 | 0.141 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| TCEAL4 |   | 1 | 1195 | 2235 | 2236 | -0.156 | 0.219 | 0.091 | undef | 0.007 |
|---|
| SH3BP4 |   | 4 | 440 | 957 | 942 | -0.105 | 0.108 | -0.100 | 0.246 | -0.003 |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
GO Enrichment output for subnetwork 6119 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.033E-09 | 2.523E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.797E-09 | 5.86E-06 |
|---|
| nucleus localization | GO:0051647 |  | 2.246E-08 | 1.829E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.59E-07 | 2.804E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.836E-07 | 2.363E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.294E-06 | 5.267E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.341E-06 | 4.681E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.348E-06 | 4.117E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.348E-06 | 3.66E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.348E-06 | 3.294E-04 |
|---|
| negative regulation of protein complex assembly | GO:0031333 |  | 1.787E-06 | 3.97E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.509E-09 | 3.631E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 7.007E-09 | 8.429E-06 |
|---|
| nucleus localization | GO:0051647 |  | 3.279E-08 | 2.63E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.432E-07 | 3.869E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.583E-07 | 3.168E-04 |
|---|
| interphase | GO:0051325 |  | 7.912E-07 | 3.173E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 8.913E-07 | 3.064E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.123E-06 | 3.377E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.123E-06 | 3.002E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.482E-06 | 3.567E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.676E-06 | 3.666E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 9.236E-10 | 2.124E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.429E-09 | 7.393E-06 |
|---|
| nucleus localization | GO:0051647 |  | 2.301E-08 | 1.764E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 2.939E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 7.146E-07 | 3.287E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 8.52E-07 | 3.266E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.52E-07 | 2.799E-04 |
|---|
| interphase | GO:0051325 |  | 9.763E-07 | 2.807E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.02E-06 | 2.606E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 2.345E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.056E-06 | 2.209E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 3.175E-08 | 5.851E-05 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 5.363E-07 | 4.942E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.363E-07 | 3.295E-04 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 7.673E-07 | 3.535E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.067E-06 | 3.933E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 1.07E-06 | 3.286E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.25E-06 | 5.924E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.105E-06 | 7.152E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.612E-06 | 7.398E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.179E-06 | 7.702E-04 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 4.458E-06 | 7.469E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 9.236E-10 | 2.124E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.429E-09 | 7.393E-06 |
|---|
| nucleus localization | GO:0051647 |  | 2.301E-08 | 1.764E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 2.939E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 7.146E-07 | 3.287E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 8.52E-07 | 3.266E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.52E-07 | 2.799E-04 |
|---|
| interphase | GO:0051325 |  | 9.763E-07 | 2.807E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.02E-06 | 2.606E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 2.345E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.056E-06 | 2.209E-04 |
|---|



