Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9616-11-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2597 | 2.300e-02 | 4.112e-02 | 8.720e-02 |
|---|
| IPC-NIBC-129 | 0.1232 | 1.972e-02 | 8.854e-03 | 6.557e-02 |
|---|
| Ivshina_GPL96-GPL97 | 0.2031 | 1.385e-01 | 1.217e-01 | 7.516e-01 |
|---|
| Loi_GPL570 | 0.3461 | 3.313e-01 | 3.467e-01 | 7.966e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1492 | 5.020e-04 | 0.000e+00 | 8.210e-04 |
|---|
| Pawitan_GPL96-GPL97 | 0.3823 | 4.260e-01 | 3.348e-01 | 9.342e-01 |
|---|
| Schmidt | 0.1090 | 2.235e-02 | 3.078e-02 | 1.253e-01 |
|---|
| Sotiriou | 0.2757 | 1.355e-01 | 1.015e-01 | 6.636e-01 |
|---|
| Zhang | 0.1126 | 2.693e-03 | 3.989e-03 | 1.305e-02 |
|---|
Expression data for subnetwork 9616-11-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
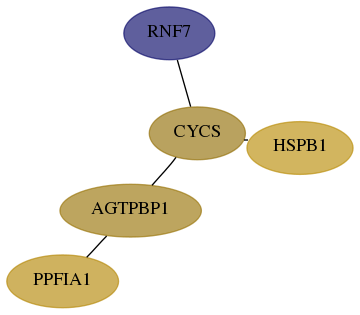
Score for each gene in subnetwork 9616-11-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| rnf7 |   | 1 | 57 | 165 | 167 | -0.027 | -0.106 | 0.061 | 0.093 | 0.074 | 0.162 | 0.083 | 0.035 | 0.257 |
|---|
| agtpbp1 |   | 2 | 35 | 118 | 111 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.026 | 0.110 | 0.151 | 0.064 |
|---|
| hspb1 |   | 5 | 13 | 118 | 99 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | 0.289 | -0.021 | 0.201 | -0.077 |
|---|
| cycs |   | 4 | 19 | 118 | 103 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | 0.272 | 0.157 | 0.077 | 0.135 |
|---|
| ppfia1 |   | 2 | 35 | 165 | 157 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.225 | 0.036 | 0.176 | 0.127 |
|---|
GO Enrichment output for subnetwork 9616-11-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.831E-06 | 4.212E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.746E-06 | 3.158E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.918E-05 | 0.01470735 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.121E-05 | 0.01794753 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.121E-05 | 0.01435803 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.831E-05 | 0.01468716 |
|---|
| response to heat | GO:0009408 |  | 7.92E-05 | 0.02602353 |
|---|
| nucleus organization | GO:0006997 |  | 9.607E-05 | 0.0276198 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.417E-04 | 0.03621035 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.563E-04 | 0.03595711 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.639E-04 | 0.03427499 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 2.204E-10 | 5.385E-07 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 7.712E-10 | 9.42E-07 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.234E-09 | 1.005E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.496E-08 | 9.135E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.924E-08 | 1.428E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.385E-08 | 1.378E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.447E-08 | 1.552E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.165E-07 | 6.611E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.107E-07 | 8.432E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.785E-07 | 9.246E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.287E-07 | 9.522E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 4.83E-10 | 1.162E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.69E-09 | 2.033E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.703E-09 | 2.168E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.276E-08 | 1.97E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.401E-08 | 3.08E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.411E-08 | 2.972E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.735E-08 | 3.346E-05 |
|---|
| nucleus organization | GO:0006997 |  | 4.045E-07 | 1.217E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.793E-07 | 1.816E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 8.275E-07 | 1.991E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.275E-07 | 1.81E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 2.204E-10 | 5.385E-07 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 7.712E-10 | 9.42E-07 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.234E-09 | 1.005E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.496E-08 | 9.135E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.924E-08 | 1.428E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.385E-08 | 1.378E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.447E-08 | 1.552E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.165E-07 | 6.611E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.107E-07 | 8.432E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.785E-07 | 9.246E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.287E-07 | 9.522E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 4.83E-10 | 1.162E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.69E-09 | 2.033E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.703E-09 | 2.168E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.276E-08 | 1.97E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.401E-08 | 3.08E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.411E-08 | 2.972E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.735E-08 | 3.346E-05 |
|---|
| nucleus organization | GO:0006997 |  | 4.045E-07 | 1.217E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.793E-07 | 1.816E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 8.275E-07 | 1.991E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.275E-07 | 1.81E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 4.83E-10 | 1.162E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.69E-09 | 2.033E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.703E-09 | 2.168E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.276E-08 | 1.97E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.401E-08 | 3.08E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.411E-08 | 2.972E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.735E-08 | 3.346E-05 |
|---|
| nucleus organization | GO:0006997 |  | 4.045E-07 | 1.217E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.793E-07 | 1.816E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 8.275E-07 | 1.991E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.275E-07 | 1.81E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.831E-06 | 4.212E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.746E-06 | 3.158E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.918E-05 | 0.01470735 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.121E-05 | 0.01794753 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.121E-05 | 0.01435803 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.831E-05 | 0.01468716 |
|---|
| response to heat | GO:0009408 |  | 7.92E-05 | 0.02602353 |
|---|
| nucleus organization | GO:0006997 |  | 9.607E-05 | 0.0276198 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.417E-04 | 0.03621035 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.563E-04 | 0.03595711 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.639E-04 | 0.03427499 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.831E-06 | 4.212E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.746E-06 | 3.158E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.918E-05 | 0.01470735 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.121E-05 | 0.01794753 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.121E-05 | 0.01435803 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.831E-05 | 0.01468716 |
|---|
| response to heat | GO:0009408 |  | 7.92E-05 | 0.02602353 |
|---|
| nucleus organization | GO:0006997 |  | 9.607E-05 | 0.0276198 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.417E-04 | 0.03621035 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.563E-04 | 0.03595711 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.639E-04 | 0.03427499 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.831E-06 | 4.212E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.746E-06 | 3.158E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.918E-05 | 0.01470735 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.121E-05 | 0.01794753 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.121E-05 | 0.01435803 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.831E-05 | 0.01468716 |
|---|
| response to heat | GO:0009408 |  | 7.92E-05 | 0.02602353 |
|---|
| nucleus organization | GO:0006997 |  | 9.607E-05 | 0.0276198 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.417E-04 | 0.03621035 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.563E-04 | 0.03595711 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.639E-04 | 0.03427499 |
|---|



