Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9276-11-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1848 | 1.222e-02 | 2.341e-02 | 4.905e-02 |
|---|
| IPC-NIBC-129 | 0.2195 | 4.848e-02 | 2.599e-02 | 1.954e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2336 | 2.074e-02 | 1.280e-02 | 2.010e-01 |
|---|
| Loi_GPL570 | 0.2753 | 1.039e-01 | 7.893e-02 | 4.026e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2132 | 8.336e-03 | 5.300e-05 | 8.058e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3049 | 1.066e-01 | 5.053e-02 | 5.013e-01 |
|---|
| Schmidt | 0.1963 | 1.798e-02 | 2.495e-02 | 1.011e-01 |
|---|
| Sotiriou | 0.2894 | 4.641e-02 | 3.295e-02 | 3.206e-01 |
|---|
| Zhang | 0.2470 | 9.809e-03 | 1.458e-02 | 3.996e-02 |
|---|
Expression data for subnetwork 9276-11-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
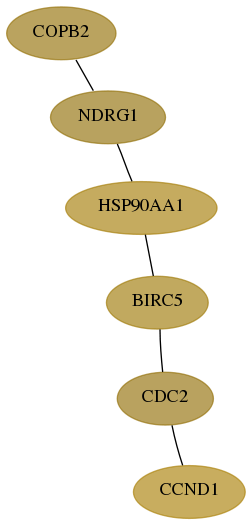
Score for each gene in subnetwork 9276-11-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| copb2 |   | 1 | 57 | 37 | 46 | 0.076 | 0.119 | 0.040 | -0.111 | 0.053 | 0.163 | 0.122 | -0.019 | 0.068 |
|---|
| ndrg1 |   | 3 | 25 | 37 | 34 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.083 | 0.068 | 0.064 | 0.092 |
|---|
| birc5 |   | 8 | 11 | 37 | 28 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.202 | 0.236 | 0.241 | 0.205 |
|---|
| hsp90aa1 |   | 6 | 12 | 37 | 29 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.169 | 0.069 | 0.102 | -0.011 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 9276-11-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.951E-06 | 7.994E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 5.861E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 4.395E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 3.516E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.101E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 3.515E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.075E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 2.734E-03 |
|---|
| organelle localization | GO:0051640 |  | 1.185E-05 | 2.724E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 2.981E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.04E-06 | 2.54E-03 |
|---|
| establishment of organelle localization | GO:0051656 |  | 1.949E-06 | 2.38E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.491E-06 | 2.028E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.486E-06 | 2.129E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.486E-06 | 1.703E-03 |
|---|
| organelle localization | GO:0051640 |  | 3.871E-06 | 1.576E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.648E-06 | 1.622E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.648E-06 | 1.419E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.648E-06 | 1.262E-03 |
|---|
| mast cell activation | GO:0045576 |  | 4.648E-06 | 1.135E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.648E-06 | 1.032E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.706E-06 | 4.104E-03 |
|---|
| establishment of organelle localization | GO:0051656 |  | 3.195E-06 | 3.844E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.469E-06 | 2.782E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.855E-06 | 2.92E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 4.855E-06 | 2.336E-03 |
|---|
| organelle localization | GO:0051640 |  | 6.343E-06 | 2.544E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.472E-06 | 2.224E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.472E-06 | 1.946E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 6.472E-06 | 1.73E-03 |
|---|
| mast cell activation | GO:0045576 |  | 6.472E-06 | 1.557E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.472E-06 | 1.416E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.04E-06 | 2.54E-03 |
|---|
| establishment of organelle localization | GO:0051656 |  | 1.949E-06 | 2.38E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.491E-06 | 2.028E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.486E-06 | 2.129E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.486E-06 | 1.703E-03 |
|---|
| organelle localization | GO:0051640 |  | 3.871E-06 | 1.576E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.648E-06 | 1.622E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.648E-06 | 1.419E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.648E-06 | 1.262E-03 |
|---|
| mast cell activation | GO:0045576 |  | 4.648E-06 | 1.135E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.648E-06 | 1.032E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.706E-06 | 4.104E-03 |
|---|
| establishment of organelle localization | GO:0051656 |  | 3.195E-06 | 3.844E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.469E-06 | 2.782E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.855E-06 | 2.92E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 4.855E-06 | 2.336E-03 |
|---|
| organelle localization | GO:0051640 |  | 6.343E-06 | 2.544E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.472E-06 | 2.224E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.472E-06 | 1.946E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 6.472E-06 | 1.73E-03 |
|---|
| mast cell activation | GO:0045576 |  | 6.472E-06 | 1.557E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.472E-06 | 1.416E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.706E-06 | 4.104E-03 |
|---|
| establishment of organelle localization | GO:0051656 |  | 3.195E-06 | 3.844E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.469E-06 | 2.782E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.855E-06 | 2.92E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 4.855E-06 | 2.336E-03 |
|---|
| organelle localization | GO:0051640 |  | 6.343E-06 | 2.544E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.472E-06 | 2.224E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.472E-06 | 1.946E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 6.472E-06 | 1.73E-03 |
|---|
| mast cell activation | GO:0045576 |  | 6.472E-06 | 1.557E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.472E-06 | 1.416E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.951E-06 | 7.994E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 5.861E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 4.395E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 3.516E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.101E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 3.515E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.075E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 2.734E-03 |
|---|
| organelle localization | GO:0051640 |  | 1.185E-05 | 2.724E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 2.981E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.951E-06 | 7.994E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 5.861E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 4.395E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 3.516E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.101E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 3.515E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.075E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 2.734E-03 |
|---|
| organelle localization | GO:0051640 |  | 1.185E-05 | 2.724E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 2.981E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.951E-06 | 7.994E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 5.861E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 4.395E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 3.516E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.101E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 3.515E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.075E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 2.734E-03 |
|---|
| organelle localization | GO:0051640 |  | 1.185E-05 | 2.724E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 2.981E-03 |
|---|



