Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7332-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1763 | 1.150e-02 | 2.219e-02 | 4.640e-02 |
|---|
| IPC-NIBC-129 | 0.1459 | 1.050e-01 | 6.562e-02 | 4.243e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2365 | 8.949e-02 | 7.295e-02 | 6.046e-01 |
|---|
| Loi_GPL570 | 0.2132 | 2.573e-01 | 2.544e-01 | 7.073e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2277 | 1.132e-02 | 1.310e-04 | 1.197e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3236 | 1.114e-01 | 5.367e-02 | 5.156e-01 |
|---|
| Schmidt | 0.1287 | 1.941e-02 | 2.686e-02 | 1.091e-01 |
|---|
| Sotiriou | 0.2846 | 3.625e-02 | 2.548e-02 | 2.608e-01 |
|---|
| Zhang | 0.2430 | 7.189e-03 | 1.069e-02 | 3.063e-02 |
|---|
Expression data for subnetwork 7332-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
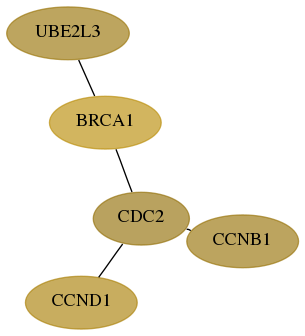
Score for each gene in subnetwork 7332-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| ube2l3 |   | 1 | 57 | 159 | 163 | 0.086 | 0.083 | 0.121 | 0.073 | 0.088 | 0.097 | 0.078 | -0.001 | 0.053 |
|---|
| ccnb1 |   | 25 | 3 | 46 | 33 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.351 | 0.129 | 0.249 | 0.167 |
|---|
| brca1 |   | 3 | 25 | 111 | 98 | 0.192 | 0.052 | 0.112 | 0.022 | 0.125 | 0.113 | 0.077 | 0.168 | 0.064 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 7332-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 4.314E-06 | 9.923E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 6.469E-06 | 7.439E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 6.469E-06 | 4.96E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 6.469E-06 | 3.72E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.696E-06 | 4E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 3.47E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 9.053E-06 | 2.975E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.135E-06 | 2.626E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.054E-05 | 2.693E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.054E-05 | 2.424E-03 |
|---|
| regulation of transcription from RNA polymerase III promoter | GO:0006359 |  | 1.207E-05 | 2.523E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.234E-06 | 3.015E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.851E-06 | 2.261E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.851E-06 | 1.507E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.889E-06 | 1.154E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.975E-06 | 9.648E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.246E-06 | 9.146E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.246E-06 | 7.84E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.59E-06 | 7.911E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.752E-06 | 7.471E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.974E-06 | 7.266E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.453E-06 | 7.669E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.719E-06 | 4.135E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.577E-06 | 3.101E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.577E-06 | 2.067E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.098E-06 | 1.863E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.238E-06 | 1.558E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.607E-06 | 1.447E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.683E-06 | 1.266E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.683E-06 | 1.108E-03 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 4.512E-06 | 1.206E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 1.157E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.875E-06 | 1.066E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.234E-06 | 3.015E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.851E-06 | 2.261E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.851E-06 | 1.507E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.889E-06 | 1.154E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.975E-06 | 9.648E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.246E-06 | 9.146E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.246E-06 | 7.84E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.59E-06 | 7.911E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.752E-06 | 7.471E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.974E-06 | 7.266E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.453E-06 | 7.669E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.719E-06 | 4.135E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.577E-06 | 3.101E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.577E-06 | 2.067E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.098E-06 | 1.863E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.238E-06 | 1.558E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.607E-06 | 1.447E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.683E-06 | 1.266E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.683E-06 | 1.108E-03 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 4.512E-06 | 1.206E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 1.157E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.875E-06 | 1.066E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.719E-06 | 4.135E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.577E-06 | 3.101E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.577E-06 | 2.067E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.098E-06 | 1.863E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.238E-06 | 1.558E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.607E-06 | 1.447E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.683E-06 | 1.266E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.683E-06 | 1.108E-03 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 4.512E-06 | 1.206E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 1.157E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.875E-06 | 1.066E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 4.314E-06 | 9.923E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 6.469E-06 | 7.439E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 6.469E-06 | 4.96E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 6.469E-06 | 3.72E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.696E-06 | 4E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 3.47E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 9.053E-06 | 2.975E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.135E-06 | 2.626E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.054E-05 | 2.693E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.054E-05 | 2.424E-03 |
|---|
| regulation of transcription from RNA polymerase III promoter | GO:0006359 |  | 1.207E-05 | 2.523E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 4.314E-06 | 9.923E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 6.469E-06 | 7.439E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 6.469E-06 | 4.96E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 6.469E-06 | 3.72E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.696E-06 | 4E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 3.47E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 9.053E-06 | 2.975E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.135E-06 | 2.626E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.054E-05 | 2.693E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.054E-05 | 2.424E-03 |
|---|
| regulation of transcription from RNA polymerase III promoter | GO:0006359 |  | 1.207E-05 | 2.523E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 4.314E-06 | 9.923E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 6.469E-06 | 7.439E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 6.469E-06 | 4.96E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 6.469E-06 | 3.72E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.696E-06 | 4E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 3.47E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 9.053E-06 | 2.975E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.135E-06 | 2.626E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.054E-05 | 2.693E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.054E-05 | 2.424E-03 |
|---|
| regulation of transcription from RNA polymerase III promoter | GO:0006359 |  | 1.207E-05 | 2.523E-03 |
|---|



