Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6949-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1714 | 1.585e-02 | 2.953e-02 | 6.231e-02 |
|---|
| IPC-NIBC-129 | 0.1985 | 3.624e-02 | 1.834e-02 | 1.398e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2211 | 3.163e-02 | 2.122e-02 | 2.903e-01 |
|---|
| Loi_GPL570 | 0.2983 | 7.841e-02 | 5.418e-02 | 3.269e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1745 | 1.348e-02 | 2.180e-04 | 1.484e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3037 | 1.224e-01 | 6.106e-02 | 5.468e-01 |
|---|
| Schmidt | 0.2167 | 1.876e-02 | 2.600e-02 | 1.055e-01 |
|---|
| Sotiriou | 0.2867 | 8.921e-02 | 6.528e-02 | 5.182e-01 |
|---|
| Zhang | 0.2342 | 9.999e-03 | 1.486e-02 | 4.061e-02 |
|---|
Expression data for subnetwork 6949-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
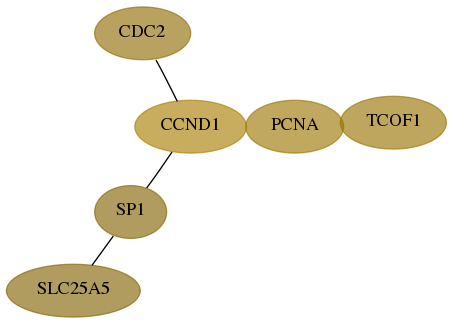
Score for each gene in subnetwork 6949-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| sp1 |   | 14 | 4 | 11 | 6 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.091 |
|---|
| pcna |   | 9 | 7 | 56 | 42 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.123 |
|---|
| tcof1 |   | 1 | 57 | 76 | 80 | 0.089 | 0.213 | 0.113 | 0.106 | 0.088 | -0.033 | 0.173 | 0.106 | 0.022 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| slc25a5 |   | 5 | 13 | 29 | 20 | 0.053 | 0.214 | 0.106 | 0.218 | 0.115 | 0.221 | 0.177 | 0.141 | -0.019 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 6949-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 3.596E-06 | 4.135E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 5.392E-06 | 4.134E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 4.339E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 7.546E-06 | 3.471E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 1.006E-05 | 3.855E-03 |
|---|
| zymogen activation | GO:0031638 |  | 1.615E-05 | 5.307E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.974E-05 | 5.674E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 9.615E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 9.887E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.299E-05 | 8.988E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.041E-07 | 1.232E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.044E-06 | 1.276E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.566E-06 | 1.275E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.566E-06 | 9.565E-04 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 2.192E-06 | 1.071E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 2.192E-06 | 8.926E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 1.02E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 4.695E-06 | 1.434E-03 |
|---|
| zymogen activation | GO:0031638 |  | 6.883E-06 | 1.868E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 9.487E-06 | 2.318E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.251E-05 | 2.777E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 1.991E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.454E-06 | 1.75E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.454E-06 | 1.166E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.181E-06 | 1.312E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 2.181E-06 | 1.05E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 3.053E-06 | 1.224E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.399E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 5.231E-06 | 1.573E-03 |
|---|
| zymogen activation | GO:0031638 |  | 7.988E-06 | 2.136E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.321E-05 | 3.178E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.741E-05 | 3.808E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.041E-07 | 1.232E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.044E-06 | 1.276E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.566E-06 | 1.275E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.566E-06 | 9.565E-04 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 2.192E-06 | 1.071E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 2.192E-06 | 8.926E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 1.02E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 4.695E-06 | 1.434E-03 |
|---|
| zymogen activation | GO:0031638 |  | 6.883E-06 | 1.868E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 9.487E-06 | 2.318E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.251E-05 | 2.777E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 1.991E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.454E-06 | 1.75E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.454E-06 | 1.166E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.181E-06 | 1.312E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 2.181E-06 | 1.05E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 3.053E-06 | 1.224E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.399E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 5.231E-06 | 1.573E-03 |
|---|
| zymogen activation | GO:0031638 |  | 7.988E-06 | 2.136E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.321E-05 | 3.178E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.741E-05 | 3.808E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 1.991E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.454E-06 | 1.75E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.454E-06 | 1.166E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.181E-06 | 1.312E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 2.181E-06 | 1.05E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 3.053E-06 | 1.224E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.399E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 5.231E-06 | 1.573E-03 |
|---|
| zymogen activation | GO:0031638 |  | 7.988E-06 | 2.136E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.321E-05 | 3.178E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.741E-05 | 3.808E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 3.596E-06 | 4.135E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 5.392E-06 | 4.134E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 4.339E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 7.546E-06 | 3.471E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 1.006E-05 | 3.855E-03 |
|---|
| zymogen activation | GO:0031638 |  | 1.615E-05 | 5.307E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.974E-05 | 5.674E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 9.615E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 9.887E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.299E-05 | 8.988E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 3.596E-06 | 4.135E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 5.392E-06 | 4.134E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 4.339E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 7.546E-06 | 3.471E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 1.006E-05 | 3.855E-03 |
|---|
| zymogen activation | GO:0031638 |  | 1.615E-05 | 5.307E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.974E-05 | 5.674E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 9.615E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 9.887E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.299E-05 | 8.988E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 3.596E-06 | 4.135E-03 |
|---|
| positive regulation of fibrinolysis | GO:0051919 |  | 5.392E-06 | 4.134E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 4.339E-03 |
|---|
| transcription from RNA polymerase I promoter | GO:0006360 |  | 7.546E-06 | 3.471E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 1.006E-05 | 3.855E-03 |
|---|
| zymogen activation | GO:0031638 |  | 1.615E-05 | 5.307E-03 |
|---|
| rRNA transcription | GO:0009303 |  | 1.974E-05 | 5.674E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 9.615E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 9.887E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.299E-05 | 8.988E-03 |
|---|



