Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6502-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1162 | 1.215e-02 | 2.331e-02 | 4.883e-02 |
|---|
| IPC-NIBC-129 | 0.1660 | 7.815e-02 | 4.606e-02 | 3.223e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2339 | 6.026e-02 | 4.572e-02 | 4.727e-01 |
|---|
| Loi_GPL570 | 0.2371 | 8.023e-02 | 5.587e-02 | 3.327e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1690 | 8.862e-02 | 3.490e-02 | 7.490e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3476 | 6.900e-02 | 2.777e-02 | 3.710e-01 |
|---|
| Schmidt | 0.2151 | 2.435e-02 | 3.342e-02 | 1.361e-01 |
|---|
| Sotiriou | 0.2703 | 9.787e-02 | 7.196e-02 | 5.498e-01 |
|---|
| Zhang | 0.2870 | 4.818e-03 | 7.158e-03 | 2.168e-02 |
|---|
Expression data for subnetwork 6502-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
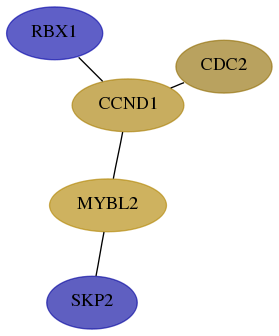
Score for each gene in subnetwork 6502-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| rbx1 |   | 1 | 57 | 123 | 124 | -0.125 | 0.081 | 0.140 | 0.166 | 0.088 | 0.169 | 0.155 | -0.043 | 0.154 |
|---|
| mybl2 |   | 9 | 7 | 29 | 13 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.275 | 0.202 | 0.182 | 0.074 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| skp2 |   | 1 | 57 | 123 | 124 | -0.101 | 0.121 | 0.060 | 0.145 | 0.073 | 0.188 | 0.224 | 0.031 | 0.000 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 6502-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.643E-06 | 3.78E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 0.01031444 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 8.29E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.01618709 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02357018 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02432496 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.0232122 |
|---|
| response to calcium ion | GO:0051592 |  | 2.271E-04 | 0.05802679 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.878E-04 | 0.06618337 |
|---|
| response to UV | GO:0009411 |  | 3.007E-04 | 0.06288236 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.419E-07 | 5.909E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 2.434E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.405E-06 | 1.958E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 8.245E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 0.01127184 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.682E-05 | 0.01092173 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.881E-05 | 0.01005342 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.981E-05 | 0.02437062 |
|---|
| response to calcium ion | GO:0051592 |  | 8.319E-05 | 0.0225812 |
|---|
| response to UV | GO:0009411 |  | 1.05E-04 | 0.02563998 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.128E-04 | 0.02504385 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.972E-07 | 9.555E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 3.339E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.59E-06 | 2.88E-03 |
|---|
| interphase | GO:0051325 |  | 3.943E-06 | 2.372E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 8.14E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 0.01188858 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 0.01191832 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.734E-05 | 0.01122874 |
|---|
| response to calcium ion | GO:0051592 |  | 1.064E-04 | 0.02844793 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.11E-04 | 0.02671159 |
|---|
| response to UV | GO:0009411 |  | 1.355E-04 | 0.02963893 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.419E-07 | 5.909E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 2.434E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.405E-06 | 1.958E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 8.245E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 0.01127184 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.682E-05 | 0.01092173 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.881E-05 | 0.01005342 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.981E-05 | 0.02437062 |
|---|
| response to calcium ion | GO:0051592 |  | 8.319E-05 | 0.0225812 |
|---|
| response to UV | GO:0009411 |  | 1.05E-04 | 0.02563998 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.128E-04 | 0.02504385 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.972E-07 | 9.555E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 3.339E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.59E-06 | 2.88E-03 |
|---|
| interphase | GO:0051325 |  | 3.943E-06 | 2.372E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 8.14E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 0.01188858 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 0.01191832 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.734E-05 | 0.01122874 |
|---|
| response to calcium ion | GO:0051592 |  | 1.064E-04 | 0.02844793 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.11E-04 | 0.02671159 |
|---|
| response to UV | GO:0009411 |  | 1.355E-04 | 0.02963893 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.972E-07 | 9.555E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 3.339E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.59E-06 | 2.88E-03 |
|---|
| interphase | GO:0051325 |  | 3.943E-06 | 2.372E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 8.14E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 0.01188858 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 0.01191832 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.734E-05 | 0.01122874 |
|---|
| response to calcium ion | GO:0051592 |  | 1.064E-04 | 0.02844793 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.11E-04 | 0.02671159 |
|---|
| response to UV | GO:0009411 |  | 1.355E-04 | 0.02963893 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.643E-06 | 3.78E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 0.01031444 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 8.29E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.01618709 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02357018 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02432496 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.0232122 |
|---|
| response to calcium ion | GO:0051592 |  | 2.271E-04 | 0.05802679 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.878E-04 | 0.06618337 |
|---|
| response to UV | GO:0009411 |  | 3.007E-04 | 0.06288236 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.643E-06 | 3.78E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 0.01031444 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 8.29E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.01618709 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02357018 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02432496 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.0232122 |
|---|
| response to calcium ion | GO:0051592 |  | 2.271E-04 | 0.05802679 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.878E-04 | 0.06618337 |
|---|
| response to UV | GO:0009411 |  | 3.007E-04 | 0.06288236 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.643E-06 | 3.78E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 0.01031444 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 8.29E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.01618709 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02357018 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02432496 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.0232122 |
|---|
| response to calcium ion | GO:0051592 |  | 2.271E-04 | 0.05802679 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.878E-04 | 0.06618337 |
|---|
| response to UV | GO:0009411 |  | 3.007E-04 | 0.06288236 |
|---|



