Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 60598-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2093 | 3.074e-02 | 5.318e-02 | 1.127e-01 |
|---|
| IPC-NIBC-129 | 0.2794 | 3.580e-02 | 1.808e-02 | 1.378e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1891 | 6.132e-03 | 2.944e-03 | 5.866e-02 |
|---|
| Loi_GPL570 | 0.2993 | 4.351e-02 | 2.436e-02 | 2.038e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1611 | 3.391e-03 | 4.000e-06 | 2.192e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3439 | 1.356e-01 | 7.026e-02 | 5.814e-01 |
|---|
| Schmidt | 0.2588 | 1.548e-02 | 2.158e-02 | 8.694e-02 |
|---|
| Sotiriou | 0.2988 | 1.115e-01 | 8.258e-02 | 5.952e-01 |
|---|
| Zhang | 0.2247 | 5.120e-03 | 7.609e-03 | 2.285e-02 |
|---|
Expression data for subnetwork 60598-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
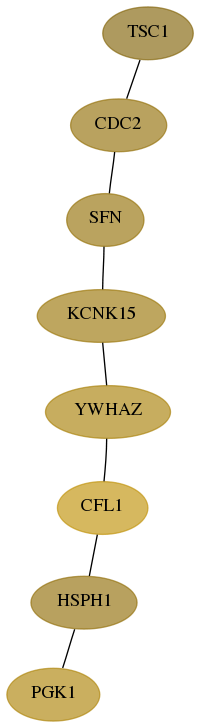
Score for each gene in subnetwork 60598-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| tsc1 |   | 11 | 6 | 1 | 3 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.196 | 0.006 | 0.010 | -0.007 |
|---|
| kcnk15 |   | 1 | 57 | 17 | 31 | 0.088 | -0.091 | -0.068 | 0.220 | 0.076 | 0.083 | 0.037 | 0.154 | 0.098 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| hsph1 |   | 2 | 35 | 17 | 17 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.161 | 0.143 | 0.132 | 0.094 |
|---|
| sfn |   | 2 | 35 | 17 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.219 | 0.100 | 0.147 | 0.079 |
|---|
| ywhaz |   | 9 | 7 | 17 | 9 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.199 | 0.056 | 0.123 | 0.103 |
|---|
| pgk1 |   | 4 | 19 | 1 | 4 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.310 | 0.201 | 0.127 | 0.054 |
|---|
| cfl1 |   | 2 | 35 | 17 | 17 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.229 | 0.174 | 0.188 | -0.025 |
|---|
GO Enrichment output for subnetwork 60598-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 3.101E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 3.902E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 9.153E-09 | 7.018E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 5.263E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 3.528E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.041E-07 | 3.989E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.504E-07 | 4.943E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.409E-07 | 6.926E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.809E-07 | 1.229E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.09E-07 | 1.171E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 7.321E-07 | 1.531E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.304E-11 | 3.187E-08 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.056E-09 | 2.512E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.056E-09 | 1.674E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 2.056E-09 | 1.256E-06 |
|---|
| neural plate development | GO:0001840 |  | 4.111E-09 | 2.008E-06 |
|---|
| glycolysis | GO:0006096 |  | 1.765E-08 | 7.186E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.385E-08 | 1.181E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.956E-08 | 1.208E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 4.511E-08 | 1.224E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 5.862E-08 | 1.432E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 8.73E-08 | 1.939E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.704E-11 | 4.1E-08 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.502E-09 | 3.01E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.502E-09 | 2.007E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.07E-08 | 1.245E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 4.118E-08 | 1.982E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.718E-08 | 1.892E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.132E-08 | 2.451E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.055E-07 | 3.173E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.395E-07 | 3.728E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.759E-07 | 4.232E-05 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.024E-07 | 1.099E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.304E-11 | 3.187E-08 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.056E-09 | 2.512E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.056E-09 | 1.674E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 2.056E-09 | 1.256E-06 |
|---|
| neural plate development | GO:0001840 |  | 4.111E-09 | 2.008E-06 |
|---|
| glycolysis | GO:0006096 |  | 1.765E-08 | 7.186E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.385E-08 | 1.181E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.956E-08 | 1.208E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 4.511E-08 | 1.224E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 5.862E-08 | 1.432E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 8.73E-08 | 1.939E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.704E-11 | 4.1E-08 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.502E-09 | 3.01E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.502E-09 | 2.007E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.07E-08 | 1.245E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 4.118E-08 | 1.982E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.718E-08 | 1.892E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.132E-08 | 2.451E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.055E-07 | 3.173E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.395E-07 | 3.728E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.759E-07 | 4.232E-05 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.024E-07 | 1.099E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.704E-11 | 4.1E-08 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.502E-09 | 3.01E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.502E-09 | 2.007E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.07E-08 | 1.245E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 4.118E-08 | 1.982E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.718E-08 | 1.892E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.132E-08 | 2.451E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.055E-07 | 3.173E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.395E-07 | 3.728E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.759E-07 | 4.232E-05 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.024E-07 | 1.099E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 3.101E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 3.902E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 9.153E-09 | 7.018E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 5.263E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 3.528E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.041E-07 | 3.989E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.504E-07 | 4.943E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.409E-07 | 6.926E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.809E-07 | 1.229E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.09E-07 | 1.171E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 7.321E-07 | 1.531E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 3.101E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 3.902E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 9.153E-09 | 7.018E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 5.263E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 3.528E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.041E-07 | 3.989E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.504E-07 | 4.943E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.409E-07 | 6.926E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.809E-07 | 1.229E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.09E-07 | 1.171E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 7.321E-07 | 1.531E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 3.101E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 3.902E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 9.153E-09 | 7.018E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 5.263E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 3.528E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.041E-07 | 3.989E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.504E-07 | 4.943E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.409E-07 | 6.926E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.809E-07 | 1.229E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.09E-07 | 1.171E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 7.321E-07 | 1.531E-04 |
|---|



