Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 55367-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2713 | 7.917e-02 | 1.224e-01 | 2.500e-01 |
|---|
| IPC-NIBC-129 | 0.2014 | 3.488e-02 | 1.752e-02 | 1.336e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1427 | 2.983e-02 | 1.978e-02 | 2.764e-01 |
|---|
| Loi_GPL570 | 0.3014 | 1.139e-01 | 8.907e-02 | 4.295e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1340 | 3.220e-04 | 0.000e+00 | 3.530e-04 |
|---|
| Pawitan_GPL96-GPL97 | 0.4414 | 4.720e-01 | 3.843e-01 | 9.532e-01 |
|---|
| Schmidt | 0.1897 | 2.810e-02 | 3.836e-02 | 1.560e-01 |
|---|
| Sotiriou | 0.2612 | 1.736e-01 | 1.321e-01 | 7.490e-01 |
|---|
| Zhang | 0.1018 | 9.970e-04 | 1.460e-03 | 5.416e-03 |
|---|
Expression data for subnetwork 55367-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
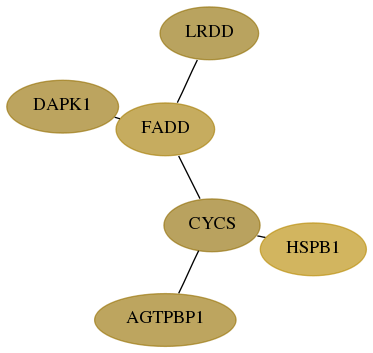
Score for each gene in subnetwork 55367-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| agtpbp1 |   | 2 | 35 | 118 | 111 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.026 | 0.110 | 0.151 | 0.064 |
|---|
| hspb1 |   | 5 | 13 | 118 | 99 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | 0.289 | -0.021 | 0.201 | -0.077 |
|---|
| cycs |   | 4 | 19 | 118 | 103 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | 0.272 | 0.157 | 0.077 | 0.135 |
|---|
| dapk1 |   | 1 | 57 | 118 | 121 | 0.082 | 0.188 | -0.078 | -0.063 | 0.067 | 0.267 | 0.117 | 0.059 | 0.142 |
|---|
| lrdd |   | 1 | 57 | 118 | 121 | 0.075 | 0.117 | -0.015 | 0.179 | 0.068 | 0.104 | 0.152 | 0.007 | 0.108 |
|---|
| fadd |   | 2 | 35 | 83 | 77 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.300 | 0.070 | 0.116 | 0.032 |
|---|
GO Enrichment output for subnetwork 55367-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.17E-07 | 1.189E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.06E-06 | 2.369E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.07E-05 | 8.201E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.439E-05 | 8.277E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.343E-05 | 0.0107755 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 9.449E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 2.876E-05 | 8.268E-03 |
|---|
| response to heat | GO:0009408 |  | 5.947E-05 | 0.01519661 |
|---|
| nucleus organization | GO:0006997 |  | 7.214E-05 | 0.01659149 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.174E-04 | 0.02455364 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.645E-09 | 4.018E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.994E-08 | 2.435E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.897E-08 | 3.173E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.511E-08 | 2.755E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 5.926E-08 | 2.896E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.884E-07 | 1.174E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 5.041E-07 | 1.759E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.711E-07 | 1.744E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.711E-07 | 1.55E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 8.514E-07 | 2.08E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.951E-07 | 2.21E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.703E-09 | 6.504E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.276E-08 | 3.941E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.401E-08 | 5.134E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.411E-08 | 4.457E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.735E-08 | 4.685E-05 |
|---|
| nucleus organization | GO:0006997 |  | 4.045E-07 | 1.622E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 8.275E-07 | 2.844E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.275E-07 | 2.489E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 8.812E-07 | 2.356E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.253E-06 | 3.015E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.551E-06 | 3.393E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.645E-09 | 4.018E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.994E-08 | 2.435E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.897E-08 | 3.173E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.511E-08 | 2.755E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 5.926E-08 | 2.896E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.884E-07 | 1.174E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 5.041E-07 | 1.759E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.711E-07 | 1.744E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.711E-07 | 1.55E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 8.514E-07 | 2.08E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.951E-07 | 2.21E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.703E-09 | 6.504E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.276E-08 | 3.941E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.401E-08 | 5.134E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.411E-08 | 4.457E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.735E-08 | 4.685E-05 |
|---|
| nucleus organization | GO:0006997 |  | 4.045E-07 | 1.622E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 8.275E-07 | 2.844E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.275E-07 | 2.489E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 8.812E-07 | 2.356E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.253E-06 | 3.015E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.551E-06 | 3.393E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.703E-09 | 6.504E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.276E-08 | 3.941E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.401E-08 | 5.134E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.411E-08 | 4.457E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.735E-08 | 4.685E-05 |
|---|
| nucleus organization | GO:0006997 |  | 4.045E-07 | 1.622E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 8.275E-07 | 2.844E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.275E-07 | 2.489E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 8.812E-07 | 2.356E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.253E-06 | 3.015E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.551E-06 | 3.393E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.17E-07 | 1.189E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.06E-06 | 2.369E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.07E-05 | 8.201E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.439E-05 | 8.277E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.343E-05 | 0.0107755 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 9.449E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 2.876E-05 | 8.268E-03 |
|---|
| response to heat | GO:0009408 |  | 5.947E-05 | 0.01519661 |
|---|
| nucleus organization | GO:0006997 |  | 7.214E-05 | 0.01659149 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.174E-04 | 0.02455364 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.17E-07 | 1.189E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.06E-06 | 2.369E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.07E-05 | 8.201E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.439E-05 | 8.277E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.343E-05 | 0.0107755 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 9.449E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 2.876E-05 | 8.268E-03 |
|---|
| response to heat | GO:0009408 |  | 5.947E-05 | 0.01519661 |
|---|
| nucleus organization | GO:0006997 |  | 7.214E-05 | 0.01659149 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.174E-04 | 0.02455364 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.17E-07 | 1.189E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.06E-06 | 2.369E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.07E-05 | 8.201E-03 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.439E-05 | 8.277E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.343E-05 | 0.0107755 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 9.449E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 2.876E-05 | 8.268E-03 |
|---|
| response to heat | GO:0009408 |  | 5.947E-05 | 0.01519661 |
|---|
| nucleus organization | GO:0006997 |  | 7.214E-05 | 0.01659149 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.174E-04 | 0.02455364 |
|---|



