Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5425-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1466 | 9.487e-03 | 1.868e-02 | 3.878e-02 |
|---|
| IPC-NIBC-129 | 0.1606 | 5.457e-02 | 2.994e-02 | 2.224e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2457 | 6.709e-02 | 5.192e-02 | 5.075e-01 |
|---|
| Loi_GPL570 | 0.2659 | 1.350e-01 | 1.114e-01 | 4.824e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2053 | 3.212e-02 | 2.527e-03 | 3.713e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3167 | 5.424e-02 | 1.993e-02 | 3.092e-01 |
|---|
| Schmidt | 0.1772 | 4.285e-02 | 5.753e-02 | 2.294e-01 |
|---|
| Sotiriou | 0.2343 | 5.310e-02 | 3.791e-02 | 3.568e-01 |
|---|
| Zhang | 0.3089 | 8.054e-03 | 1.197e-02 | 3.376e-02 |
|---|
Expression data for subnetwork 5425-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
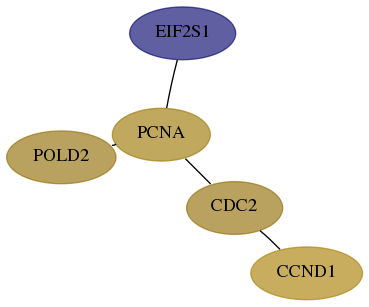
Score for each gene in subnetwork 5425-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| pold2 |   | 3 | 25 | 101 | 88 | 0.073 | 0.180 | 0.204 | 0.131 | 0.150 | 0.186 | 0.146 | 0.126 | 0.072 |
|---|
| pcna |   | 9 | 7 | 56 | 42 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.123 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| eif2s1 |   | 2 | 35 | 105 | 96 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.239 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 5425-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 4.631E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.02471707 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.03454455 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.03465783 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.86E-05 | 0.0323962 |
|---|
| base-excision repair | GO:0006284 |  | 9.86E-05 | 0.02834667 |
|---|
| response to calcium ion | GO:0051592 |  | 2.771E-04 | 0.07081896 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.205E-04 | 0.07371195 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.511E-04 | 0.07341664 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.232E-08 | 3.01E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 6.393E-07 | 7.809E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 1.983E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.649E-05 | 0.01007325 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.818E-05 | 0.01377049 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.277E-05 | 0.01334228 |
|---|
| base-excision repair | GO:0006284 |  | 3.277E-05 | 0.01143624 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.519E-05 | 0.01074613 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.945E-05 | 0.02428105 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.746E-05 | 0.0238085 |
|---|
| response to calcium ion | GO:0051592 |  | 1.016E-04 | 0.02256131 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.024E-08 | 4.871E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 9.941E-07 | 1.196E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.392E-06 | 2.72E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.067E-05 | 0.0124321 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.622E-05 | 0.01742771 |
|---|
| fat cell differentiation | GO:0045444 |  | 4.236E-05 | 0.01698524 |
|---|
| base-excision repair | GO:0006284 |  | 4.236E-05 | 0.01455878 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 4.561E-05 | 0.01371612 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.244E-04 | 0.03326363 |
|---|
| response to calcium ion | GO:0051592 |  | 1.299E-04 | 0.03126158 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.356E-04 | 0.02964934 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.232E-08 | 3.01E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 6.393E-07 | 7.809E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 1.983E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.649E-05 | 0.01007325 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.818E-05 | 0.01377049 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.277E-05 | 0.01334228 |
|---|
| base-excision repair | GO:0006284 |  | 3.277E-05 | 0.01143624 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.519E-05 | 0.01074613 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.945E-05 | 0.02428105 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.746E-05 | 0.0238085 |
|---|
| response to calcium ion | GO:0051592 |  | 1.016E-04 | 0.02256131 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.024E-08 | 4.871E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 9.941E-07 | 1.196E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.392E-06 | 2.72E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.067E-05 | 0.0124321 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.622E-05 | 0.01742771 |
|---|
| fat cell differentiation | GO:0045444 |  | 4.236E-05 | 0.01698524 |
|---|
| base-excision repair | GO:0006284 |  | 4.236E-05 | 0.01455878 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 4.561E-05 | 0.01371612 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.244E-04 | 0.03326363 |
|---|
| response to calcium ion | GO:0051592 |  | 1.299E-04 | 0.03126158 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.356E-04 | 0.02964934 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.024E-08 | 4.871E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 9.941E-07 | 1.196E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.392E-06 | 2.72E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.067E-05 | 0.0124321 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.622E-05 | 0.01742771 |
|---|
| fat cell differentiation | GO:0045444 |  | 4.236E-05 | 0.01698524 |
|---|
| base-excision repair | GO:0006284 |  | 4.236E-05 | 0.01455878 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 4.561E-05 | 0.01371612 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.244E-04 | 0.03326363 |
|---|
| response to calcium ion | GO:0051592 |  | 1.299E-04 | 0.03126158 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.356E-04 | 0.02964934 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 4.631E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.02471707 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.03454455 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.03465783 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.86E-05 | 0.0323962 |
|---|
| base-excision repair | GO:0006284 |  | 9.86E-05 | 0.02834667 |
|---|
| response to calcium ion | GO:0051592 |  | 2.771E-04 | 0.07081896 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.205E-04 | 0.07371195 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.511E-04 | 0.07341664 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 4.631E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.02471707 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.03454455 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.03465783 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.86E-05 | 0.0323962 |
|---|
| base-excision repair | GO:0006284 |  | 9.86E-05 | 0.02834667 |
|---|
| response to calcium ion | GO:0051592 |  | 2.771E-04 | 0.07081896 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.205E-04 | 0.07371195 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.511E-04 | 0.07341664 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 4.631E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.02471707 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.03454455 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.03465783 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.86E-05 | 0.0323962 |
|---|
| base-excision repair | GO:0006284 |  | 9.86E-05 | 0.02834667 |
|---|
| response to calcium ion | GO:0051592 |  | 2.771E-04 | 0.07081896 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.205E-04 | 0.07371195 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.511E-04 | 0.07341664 |
|---|



