Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5424-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1894 | 1.184e-02 | 2.277e-02 | 4.765e-02 |
|---|
| IPC-NIBC-129 | 0.1926 | 1.010e-01 | 6.262e-02 | 4.099e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2351 | 3.556e-02 | 2.441e-02 | 3.195e-01 |
|---|
| Loi_GPL570 | 0.2164 | 1.180e-01 | 9.333e-02 | 4.402e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1848 | 7.047e-03 | 3.200e-05 | 6.413e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3088 | 1.109e-01 | 5.336e-02 | 5.142e-01 |
|---|
| Schmidt | 0.1871 | 2.819e-02 | 3.848e-02 | 1.565e-01 |
|---|
| Sotiriou | 0.2610 | 7.510e-02 | 5.448e-02 | 4.612e-01 |
|---|
| Zhang | 0.2434 | 9.190e-03 | 1.366e-02 | 3.780e-02 |
|---|
Expression data for subnetwork 5424-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
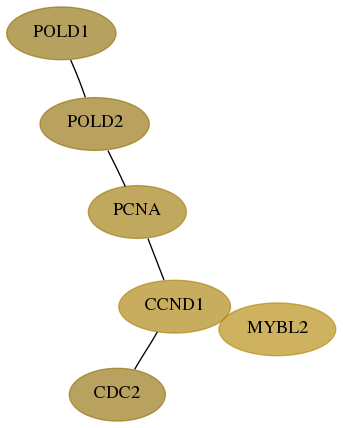
Score for each gene in subnetwork 5424-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| pold2 |   | 3 | 25 | 101 | 88 | 0.073 | 0.180 | 0.204 | 0.131 | 0.150 | 0.186 | 0.146 | 0.126 | 0.072 |
|---|
| pcna |   | 9 | 7 | 56 | 42 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.123 |
|---|
| mybl2 |   | 9 | 7 | 29 | 13 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.275 | 0.202 | 0.182 | 0.074 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| pold1 |   | 1 | 57 | 135 | 137 | 0.069 | 0.107 | 0.160 | -0.033 | 0.102 | 0.121 | 0.144 | 0.148 | 0.006 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 5424-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.792E-11 | 2.252E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.442E-08 | 1.658E-05 |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 2.078E-04 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 1.161E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 2.942E-06 | 1.353E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.367E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 2.704E-03 |
|---|
| S phase | GO:0051320 |  | 8.231E-06 | 2.366E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 3.438E-03 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 3.316E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.873E-05 | 3.916E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.739E-12 | 1.402E-08 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.239E-09 | 1.513E-06 |
|---|
| base-excision repair | GO:0006284 |  | 5.234E-08 | 4.262E-05 |
|---|
| response to UV | GO:0009411 |  | 4.175E-07 | 2.55E-04 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 7.121E-07 | 3.479E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 8.114E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.405E-06 | 8.393E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 2.562E-06 | 7.824E-04 |
|---|
| S phase | GO:0051320 |  | 2.562E-06 | 6.954E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 3.298E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 5.124E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.113E-11 | 2.678E-08 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.232E-09 | 2.685E-06 |
|---|
| base-excision repair | GO:0006284 |  | 7.678E-08 | 6.158E-05 |
|---|
| response to UV | GO:0009411 |  | 6.121E-07 | 3.682E-04 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 9.918E-07 | 4.772E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.113E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.568E-06 | 1.226E-03 |
|---|
| S phase | GO:0051320 |  | 3.568E-06 | 1.073E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.59E-06 | 9.599E-04 |
|---|
| interphase | GO:0051325 |  | 3.943E-06 | 9.488E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 3.7E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.739E-12 | 1.402E-08 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.239E-09 | 1.513E-06 |
|---|
| base-excision repair | GO:0006284 |  | 5.234E-08 | 4.262E-05 |
|---|
| response to UV | GO:0009411 |  | 4.175E-07 | 2.55E-04 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 7.121E-07 | 3.479E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 8.114E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.405E-06 | 8.393E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 2.562E-06 | 7.824E-04 |
|---|
| S phase | GO:0051320 |  | 2.562E-06 | 6.954E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 3.298E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 5.124E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.113E-11 | 2.678E-08 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.232E-09 | 2.685E-06 |
|---|
| base-excision repair | GO:0006284 |  | 7.678E-08 | 6.158E-05 |
|---|
| response to UV | GO:0009411 |  | 6.121E-07 | 3.682E-04 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 9.918E-07 | 4.772E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.113E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.568E-06 | 1.226E-03 |
|---|
| S phase | GO:0051320 |  | 3.568E-06 | 1.073E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.59E-06 | 9.599E-04 |
|---|
| interphase | GO:0051325 |  | 3.943E-06 | 9.488E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 3.7E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.113E-11 | 2.678E-08 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.232E-09 | 2.685E-06 |
|---|
| base-excision repair | GO:0006284 |  | 7.678E-08 | 6.158E-05 |
|---|
| response to UV | GO:0009411 |  | 6.121E-07 | 3.682E-04 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 9.918E-07 | 4.772E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.113E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.568E-06 | 1.226E-03 |
|---|
| S phase | GO:0051320 |  | 3.568E-06 | 1.073E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.59E-06 | 9.599E-04 |
|---|
| interphase | GO:0051325 |  | 3.943E-06 | 9.488E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 3.7E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.792E-11 | 2.252E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.442E-08 | 1.658E-05 |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 2.078E-04 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 1.161E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 2.942E-06 | 1.353E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.367E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 2.704E-03 |
|---|
| S phase | GO:0051320 |  | 8.231E-06 | 2.366E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 3.438E-03 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 3.316E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.873E-05 | 3.916E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.792E-11 | 2.252E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.442E-08 | 1.658E-05 |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 2.078E-04 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 1.161E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 2.942E-06 | 1.353E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.367E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 2.704E-03 |
|---|
| S phase | GO:0051320 |  | 8.231E-06 | 2.366E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 3.438E-03 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 3.316E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.873E-05 | 3.916E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.792E-11 | 2.252E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.442E-08 | 1.658E-05 |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 2.078E-04 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 1.161E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 2.942E-06 | 1.353E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.367E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 2.704E-03 |
|---|
| S phase | GO:0051320 |  | 8.231E-06 | 2.366E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 3.438E-03 |
|---|
| interphase | GO:0051325 |  | 1.442E-05 | 3.316E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.873E-05 | 3.916E-03 |
|---|



