Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5127-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1402 | 2.394e-02 | 4.261e-02 | 9.038e-02 |
|---|
| IPC-NIBC-129 | 0.1701 | 2.447e-02 | 1.146e-02 | 8.648e-02 |
|---|
| Ivshina_GPL96-GPL97 | 0.2012 | 5.565e-02 | 4.160e-02 | 4.475e-01 |
|---|
| Loi_GPL570 | 0.3292 | 4.742e-02 | 2.741e-02 | 2.190e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1353 | 4.004e-02 | 4.584e-03 | 4.486e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2720 | 2.181e-01 | 1.348e-01 | 7.453e-01 |
|---|
| Schmidt | 0.2527 | 1.663e-02 | 2.313e-02 | 9.349e-02 |
|---|
| Sotiriou | 0.2943 | 1.699e-01 | 1.291e-01 | 7.417e-01 |
|---|
| Zhang | 0.1795 | 1.690e-02 | 2.503e-02 | 6.329e-02 |
|---|
Expression data for subnetwork 5127-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
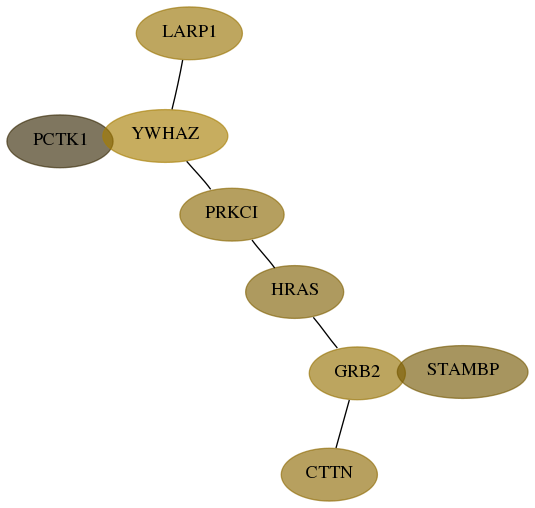
Score for each gene in subnetwork 5127-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| larp1 |   | 3 | 25 | 83 | 69 | 0.091 | 0.223 | 0.142 | 0.016 | 0.090 | 0.217 | 0.201 | 0.242 | 0.063 |
|---|
| hras |   | 3 | 25 | 58 | 50 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.245 | 0.086 | 0.208 | 0.003 |
|---|
| cttn |   | 3 | 25 | 160 | 147 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.216 | 0.069 | 0.115 | 0.025 |
|---|
| pctk1 |   | 1 | 57 | 160 | 164 | 0.008 | 0.100 | 0.114 | 0.157 | 0.107 | 0.010 | 0.044 | 0.143 | 0.128 |
|---|
| ywhaz |   | 9 | 7 | 17 | 9 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.199 | 0.056 | 0.123 | 0.103 |
|---|
| stambp |   | 2 | 35 | 160 | 152 | 0.038 | 0.150 | 0.160 | 0.131 | 0.100 | 0.223 | 0.284 | 0.065 | 0.050 |
|---|
| prkci |   | 2 | 35 | 61 | 53 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | -0.170 | 0.196 | 0.128 | 0.124 |
|---|
| grb2 |   | 3 | 25 | 160 | 147 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | 0.065 | 0.158 | 0.127 | 0.194 |
|---|
GO Enrichment output for subnetwork 5127-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.279E-08 | 2.941E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.1E-07 | 2.415E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.103E-07 | 5.445E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.442E-06 | 8.29E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.552E-06 | 1.174E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 4.591E-06 | 1.76E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 6.218E-06 | 2.043E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.491E-06 | 2.154E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.053E-05 | 2.691E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.86E-05 | 4.278E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.86E-05 | 3.889E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.702E-09 | 6.601E-06 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.701E-08 | 9.407E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.506E-07 | 1.226E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.961E-07 | 4.251E-04 |
|---|
| sperm motility | GO:0030317 |  | 8.763E-07 | 4.281E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.324E-06 | 5.392E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.903E-06 | 6.642E-04 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 3.052E-06 | 9.319E-04 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.767E-06 | 1.023E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 6.543E-06 | 1.599E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.22E-05 | 2.71E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.441E-09 | 1.069E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.265E-07 | 1.522E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.473E-07 | 1.983E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 8.9E-07 | 5.354E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.011E-06 | 4.864E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.172E-06 | 8.71E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.12E-06 | 1.072E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 4.646E-06 | 1.397E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 5.001E-06 | 1.337E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 9.111E-06 | 2.192E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.699E-05 | 3.716E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.702E-09 | 6.601E-06 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.701E-08 | 9.407E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.506E-07 | 1.226E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.961E-07 | 4.251E-04 |
|---|
| sperm motility | GO:0030317 |  | 8.763E-07 | 4.281E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.324E-06 | 5.392E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.903E-06 | 6.642E-04 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 3.052E-06 | 9.319E-04 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.767E-06 | 1.023E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 6.543E-06 | 1.599E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.22E-05 | 2.71E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.441E-09 | 1.069E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.265E-07 | 1.522E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.473E-07 | 1.983E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 8.9E-07 | 5.354E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.011E-06 | 4.864E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.172E-06 | 8.71E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.12E-06 | 1.072E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 4.646E-06 | 1.397E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 5.001E-06 | 1.337E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 9.111E-06 | 2.192E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.699E-05 | 3.716E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.441E-09 | 1.069E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.265E-07 | 1.522E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.473E-07 | 1.983E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 8.9E-07 | 5.354E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.011E-06 | 4.864E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.172E-06 | 8.71E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.12E-06 | 1.072E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 4.646E-06 | 1.397E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 5.001E-06 | 1.337E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 9.111E-06 | 2.192E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.699E-05 | 3.716E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.279E-08 | 2.941E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.1E-07 | 2.415E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.103E-07 | 5.445E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.442E-06 | 8.29E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.552E-06 | 1.174E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 4.591E-06 | 1.76E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 6.218E-06 | 2.043E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.491E-06 | 2.154E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.053E-05 | 2.691E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.86E-05 | 4.278E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.86E-05 | 3.889E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.279E-08 | 2.941E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.1E-07 | 2.415E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.103E-07 | 5.445E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.442E-06 | 8.29E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.552E-06 | 1.174E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 4.591E-06 | 1.76E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 6.218E-06 | 2.043E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.491E-06 | 2.154E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.053E-05 | 2.691E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.86E-05 | 4.278E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.86E-05 | 3.889E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.279E-08 | 2.941E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.1E-07 | 2.415E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.103E-07 | 5.445E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.442E-06 | 8.29E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.552E-06 | 1.174E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 4.591E-06 | 1.76E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 6.218E-06 | 2.043E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.491E-06 | 2.154E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.053E-05 | 2.691E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.86E-05 | 4.278E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.86E-05 | 3.889E-03 |
|---|



