Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 4869-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1461 | 1.190e-02 | 2.287e-02 | 4.787e-02 |
|---|
| IPC-NIBC-129 | 0.1681 | 5.600e-02 | 3.089e-02 | 2.288e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2349 | 5.784e-02 | 4.355e-02 | 4.596e-01 |
|---|
| Loi_GPL570 | 0.2638 | 2.623e-01 | 2.606e-01 | 7.143e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2499 | 3.267e-02 | 2.646e-03 | 3.770e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3346 | 1.026e-01 | 4.795e-02 | 4.891e-01 |
|---|
| Schmidt | 0.1272 | 1.297e-02 | 1.819e-02 | 7.254e-02 |
|---|
| Sotiriou | 0.3099 | 2.473e-02 | 1.714e-02 | 1.852e-01 |
|---|
| Zhang | 0.2506 | 5.983e-03 | 8.895e-03 | 2.615e-02 |
|---|
Expression data for subnetwork 4869-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
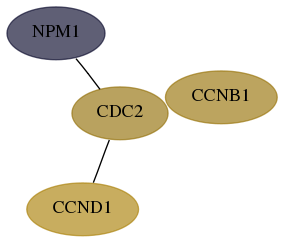
Score for each gene in subnetwork 4869-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| ccnb1 |   | 25 | 3 | 46 | 33 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.351 | 0.129 | 0.249 | 0.167 |
|---|
| npm1 |   | 1 | 57 | 93 | 95 | -0.005 | 0.172 | 0.085 | 0.197 | 0.093 | -0.050 | 0.065 | 0.169 | -0.086 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 4869-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.619E-13 | 3.724E-10 |
|---|
| ribosome assembly | GO:0042255 |  | 1.067E-11 | 1.227E-08 |
|---|
| centrosome cycle | GO:0007098 |  | 4.41E-11 | 3.381E-08 |
|---|
| cell aging | GO:0007569 |  | 1.931E-10 | 1.11E-07 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 2.359E-10 | 1.085E-07 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.817E-10 | 1.847E-07 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 8.823E-10 | 2.899E-07 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.823E-10 | 2.537E-07 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.894E-09 | 4.84E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.252E-09 | 7.48E-07 |
|---|
| positive regulation of binding | GO:0051099 |  | 3.961E-09 | 8.283E-07 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.579E-14 | 3.857E-11 |
|---|
| ribosome assembly | GO:0042255 |  | 3.157E-12 | 3.856E-09 |
|---|
| centrosome cycle | GO:0007098 |  | 9.646E-12 | 7.855E-09 |
|---|
| cell aging | GO:0007569 |  | 2.305E-11 | 1.408E-08 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 4.707E-11 | 2.3E-08 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.901E-11 | 4.031E-08 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.131E-10 | 3.949E-08 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.077E-10 | 6.341E-08 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 3.877E-10 | 1.052E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.109E-10 | 1.492E-07 |
|---|
| positive regulation of binding | GO:0051099 |  | 8.495E-10 | 1.887E-07 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 3.063E-14 | 7.37E-11 |
|---|
| ribosome assembly | GO:0042255 |  | 4.375E-12 | 5.263E-09 |
|---|
| centrosome cycle | GO:0007098 |  | 1.871E-11 | 1.5E-08 |
|---|
| cell aging | GO:0007569 |  | 3.657E-11 | 2.2E-08 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 9.128E-11 | 4.393E-08 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.45E-10 | 5.813E-08 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 2.194E-10 | 7.54E-08 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.192E-10 | 9.601E-08 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 6.817E-10 | 1.822E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 1.085E-09 | 2.612E-07 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.289E-09 | 2.819E-07 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.579E-14 | 3.857E-11 |
|---|
| ribosome assembly | GO:0042255 |  | 3.157E-12 | 3.856E-09 |
|---|
| centrosome cycle | GO:0007098 |  | 9.646E-12 | 7.855E-09 |
|---|
| cell aging | GO:0007569 |  | 2.305E-11 | 1.408E-08 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 4.707E-11 | 2.3E-08 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.901E-11 | 4.031E-08 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.131E-10 | 3.949E-08 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.077E-10 | 6.341E-08 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 3.877E-10 | 1.052E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.109E-10 | 1.492E-07 |
|---|
| positive regulation of binding | GO:0051099 |  | 8.495E-10 | 1.887E-07 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 3.063E-14 | 7.37E-11 |
|---|
| ribosome assembly | GO:0042255 |  | 4.375E-12 | 5.263E-09 |
|---|
| centrosome cycle | GO:0007098 |  | 1.871E-11 | 1.5E-08 |
|---|
| cell aging | GO:0007569 |  | 3.657E-11 | 2.2E-08 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 9.128E-11 | 4.393E-08 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.45E-10 | 5.813E-08 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 2.194E-10 | 7.54E-08 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.192E-10 | 9.601E-08 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 6.817E-10 | 1.822E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 1.085E-09 | 2.612E-07 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.289E-09 | 2.819E-07 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 3.063E-14 | 7.37E-11 |
|---|
| ribosome assembly | GO:0042255 |  | 4.375E-12 | 5.263E-09 |
|---|
| centrosome cycle | GO:0007098 |  | 1.871E-11 | 1.5E-08 |
|---|
| cell aging | GO:0007569 |  | 3.657E-11 | 2.2E-08 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 9.128E-11 | 4.393E-08 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.45E-10 | 5.813E-08 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 2.194E-10 | 7.54E-08 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.192E-10 | 9.601E-08 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 6.817E-10 | 1.822E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 1.085E-09 | 2.612E-07 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.289E-09 | 2.819E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.619E-13 | 3.724E-10 |
|---|
| ribosome assembly | GO:0042255 |  | 1.067E-11 | 1.227E-08 |
|---|
| centrosome cycle | GO:0007098 |  | 4.41E-11 | 3.381E-08 |
|---|
| cell aging | GO:0007569 |  | 1.931E-10 | 1.11E-07 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 2.359E-10 | 1.085E-07 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.817E-10 | 1.847E-07 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 8.823E-10 | 2.899E-07 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.823E-10 | 2.537E-07 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.894E-09 | 4.84E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.252E-09 | 7.48E-07 |
|---|
| positive regulation of binding | GO:0051099 |  | 3.961E-09 | 8.283E-07 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.619E-13 | 3.724E-10 |
|---|
| ribosome assembly | GO:0042255 |  | 1.067E-11 | 1.227E-08 |
|---|
| centrosome cycle | GO:0007098 |  | 4.41E-11 | 3.381E-08 |
|---|
| cell aging | GO:0007569 |  | 1.931E-10 | 1.11E-07 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 2.359E-10 | 1.085E-07 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.817E-10 | 1.847E-07 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 8.823E-10 | 2.899E-07 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.823E-10 | 2.537E-07 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.894E-09 | 4.84E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.252E-09 | 7.48E-07 |
|---|
| positive regulation of binding | GO:0051099 |  | 3.961E-09 | 8.283E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.619E-13 | 3.724E-10 |
|---|
| ribosome assembly | GO:0042255 |  | 1.067E-11 | 1.227E-08 |
|---|
| centrosome cycle | GO:0007098 |  | 4.41E-11 | 3.381E-08 |
|---|
| cell aging | GO:0007569 |  | 1.931E-10 | 1.11E-07 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 2.359E-10 | 1.085E-07 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.817E-10 | 1.847E-07 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 8.823E-10 | 2.899E-07 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.823E-10 | 2.537E-07 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.894E-09 | 4.84E-07 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.252E-09 | 7.48E-07 |
|---|
| positive regulation of binding | GO:0051099 |  | 3.961E-09 | 8.283E-07 |
|---|



