Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3735-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1928 | 8.239e-03 | 1.647e-02 | 3.398e-02 |
|---|
| IPC-NIBC-129 | 0.1674 | 3.838e-02 | 1.964e-02 | 1.496e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2525 | 5.863e-02 | 4.426e-02 | 4.639e-01 |
|---|
| Loi_GPL570 | 0.2938 | 1.570e-01 | 1.357e-01 | 5.323e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1912 | 6.232e-03 | 2.200e-05 | 5.402e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.2975 | 3.743e-02 | 1.195e-02 | 2.291e-01 |
|---|
| Schmidt | 0.1660 | 1.429e-02 | 1.999e-02 | 8.017e-02 |
|---|
| Sotiriou | 0.3038 | 6.743e-02 | 4.867e-02 | 4.272e-01 |
|---|
| Zhang | 0.3424 | 1.109e-02 | 1.647e-02 | 4.434e-02 |
|---|
Expression data for subnetwork 3735-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
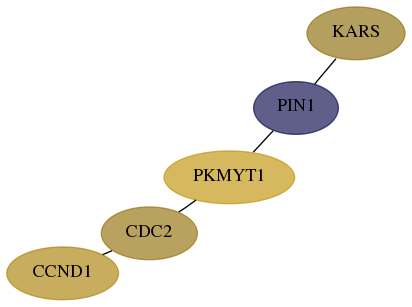
Score for each gene in subnetwork 3735-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| pin1 |   | 5 | 13 | 26 | 14 | -0.012 | 0.181 | 0.133 | 0.270 | 0.112 | 0.072 | 0.063 | 0.161 | 0.259 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| pkmyt1 |   | 5 | 13 | 26 | 14 | 0.219 | 0.222 | 0.151 | 0.190 | 0.092 | 0.202 | 0.156 | 0.061 | 0.288 |
|---|
| kars |   | 1 | 57 | 26 | 41 | 0.062 | 0.134 | 0.180 | 0.033 | 0.120 | 0.152 | 0.186 | 0.204 | 0.017 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 3735-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 4.463E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.31E-06 | 2.656E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 3.788E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 7.89E-06 | 4.536E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 0.01295659 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.923E-05 | 0.01886981 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 5.927E-05 | 0.01947556 |
|---|
| fat cell differentiation | GO:0045444 |  | 6.464E-05 | 0.01858535 |
|---|
| amino acid activation | GO:0043038 |  | 1.554E-04 | 0.03972605 |
|---|
| response to calcium ion | GO:0051592 |  | 1.819E-04 | 0.04183987 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.104E-04 | 0.04399899 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.655E-07 | 1.137E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 6.311E-07 | 7.709E-04 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 1.86E-06 | 1.514E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 1.217E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 6.596E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 9.393E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.682E-05 | 9.361E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.881E-05 | 8.797E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.325E-05 | 0.01988271 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.981E-05 | 0.01949649 |
|---|
| response to calcium ion | GO:0051592 |  | 8.319E-05 | 0.01847553 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.641E-07 | 1.838E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.036E-06 | 1.246E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 2.226E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 3.05E-06 | 1.835E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 8.14E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 0.01188858 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 0.01191832 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.734E-05 | 0.01122874 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.019E-04 | 0.02724221 |
|---|
| response to calcium ion | GO:0051592 |  | 1.064E-04 | 0.02560314 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.11E-04 | 0.02428326 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.655E-07 | 1.137E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 6.311E-07 | 7.709E-04 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 1.86E-06 | 1.514E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 1.217E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 6.596E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 9.393E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.682E-05 | 9.361E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.881E-05 | 8.797E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.325E-05 | 0.01988271 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.981E-05 | 0.01949649 |
|---|
| response to calcium ion | GO:0051592 |  | 8.319E-05 | 0.01847553 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.641E-07 | 1.838E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.036E-06 | 1.246E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 2.226E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 3.05E-06 | 1.835E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 8.14E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 0.01188858 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 0.01191832 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.734E-05 | 0.01122874 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.019E-04 | 0.02724221 |
|---|
| response to calcium ion | GO:0051592 |  | 1.064E-04 | 0.02560314 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.11E-04 | 0.02428326 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.641E-07 | 1.838E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.036E-06 | 1.246E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 2.226E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 3.05E-06 | 1.835E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 8.14E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 0.01188858 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 0.01191832 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.734E-05 | 0.01122874 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.019E-04 | 0.02724221 |
|---|
| response to calcium ion | GO:0051592 |  | 1.064E-04 | 0.02560314 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.11E-04 | 0.02428326 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 4.463E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.31E-06 | 2.656E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 3.788E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 7.89E-06 | 4.536E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 0.01295659 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.923E-05 | 0.01886981 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 5.927E-05 | 0.01947556 |
|---|
| fat cell differentiation | GO:0045444 |  | 6.464E-05 | 0.01858535 |
|---|
| amino acid activation | GO:0043038 |  | 1.554E-04 | 0.03972605 |
|---|
| response to calcium ion | GO:0051592 |  | 1.819E-04 | 0.04183987 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.104E-04 | 0.04399899 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 4.463E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.31E-06 | 2.656E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 3.788E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 7.89E-06 | 4.536E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 0.01295659 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.923E-05 | 0.01886981 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 5.927E-05 | 0.01947556 |
|---|
| fat cell differentiation | GO:0045444 |  | 6.464E-05 | 0.01858535 |
|---|
| amino acid activation | GO:0043038 |  | 1.554E-04 | 0.03972605 |
|---|
| response to calcium ion | GO:0051592 |  | 1.819E-04 | 0.04183987 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.104E-04 | 0.04399899 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 4.463E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.31E-06 | 2.656E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 3.788E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 7.89E-06 | 4.536E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 0.01295659 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.923E-05 | 0.01886981 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 5.927E-05 | 0.01947556 |
|---|
| fat cell differentiation | GO:0045444 |  | 6.464E-05 | 0.01858535 |
|---|
| amino acid activation | GO:0043038 |  | 1.554E-04 | 0.03972605 |
|---|
| response to calcium ion | GO:0051592 |  | 1.819E-04 | 0.04183987 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.104E-04 | 0.04399899 |
|---|



