Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3364-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1449 | 1.124e-02 | 2.173e-02 | 4.540e-02 |
|---|
| IPC-NIBC-129 | 0.2237 | 1.144e-01 | 7.278e-02 | 4.572e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2376 | 1.904e-02 | 1.155e-02 | 1.857e-01 |
|---|
| Loi_GPL570 | 0.2062 | 1.222e-01 | 9.775e-02 | 4.510e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1848 | 3.407e-02 | 2.967e-03 | 3.913e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3412 | 5.832e-02 | 2.202e-02 | 3.269e-01 |
|---|
| Schmidt | 0.1845 | 3.432e-02 | 4.650e-02 | 1.879e-01 |
|---|
| Sotiriou | 0.2484 | 7.516e-02 | 5.453e-02 | 4.615e-01 |
|---|
| Zhang | 0.3024 | 5.354e-03 | 7.957e-03 | 2.375e-02 |
|---|
Expression data for subnetwork 3364-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
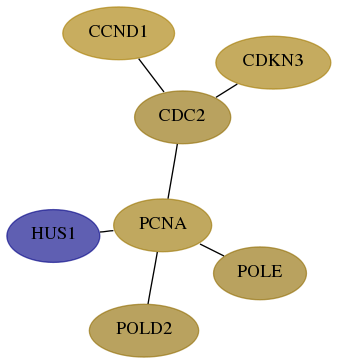
Score for each gene in subnetwork 3364-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cdkn3 |   | 12 | 5 | 31 | 16 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.323 | 0.186 | 0.198 | 0.271 |
|---|
| pold2 |   | 3 | 25 | 101 | 88 | 0.073 | 0.180 | 0.204 | 0.131 | 0.150 | 0.186 | 0.146 | 0.126 | 0.072 |
|---|
| pcna |   | 9 | 7 | 56 | 42 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.123 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| pole |   | 1 | 57 | 101 | 106 | 0.073 | 0.135 | 0.081 | -0.205 | 0.081 | 0.139 | 0.145 | 0.105 | -0.014 |
|---|
| hus1 |   | 1 | 57 | 101 | 106 | -0.058 | 0.197 | 0.116 | 0.109 | 0.101 | 0.160 | 0.072 | 0.082 | 0.071 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 3364-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.328E-10 | 7.655E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.232E-08 | 2.567E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.878E-08 | 3.74E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.773E-07 | 2.169E-04 |
|---|
| interphase | GO:0051325 |  | 4.14E-07 | 1.904E-04 |
|---|
| response to UV | GO:0009411 |  | 4.787E-06 | 1.835E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.45E-06 | 1.791E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.553E-06 | 1.884E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 2.734E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.426E-05 | 3.279E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.797E-05 | 5.848E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.725E-11 | 9.1E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.315E-09 | 4.05E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 8.003E-09 | 6.517E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.245E-08 | 4.425E-05 |
|---|
| response to UV | GO:0009411 |  | 1.574E-06 | 7.692E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.663E-06 | 6.771E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.755E-06 | 6.124E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.648E-06 | 1.419E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 5.974E-06 | 1.622E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.753E-06 | 2.138E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.293E-05 | 2.872E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.786E-11 | 9.109E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.37E-09 | 4.055E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.568E-09 | 6.069E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.498E-08 | 3.909E-05 |
|---|
| interphase | GO:0051325 |  | 7.368E-08 | 3.546E-05 |
|---|
| response to UV | GO:0009411 |  | 1.453E-06 | 5.829E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.719E-06 | 5.907E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.814E-06 | 5.455E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 1.286E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 6.181E-06 | 1.487E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 9.048E-06 | 1.979E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.725E-11 | 9.1E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.315E-09 | 4.05E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 8.003E-09 | 6.517E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.245E-08 | 4.425E-05 |
|---|
| response to UV | GO:0009411 |  | 1.574E-06 | 7.692E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.663E-06 | 6.771E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.755E-06 | 6.124E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.648E-06 | 1.419E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 5.974E-06 | 1.622E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.753E-06 | 2.138E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.293E-05 | 2.872E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.786E-11 | 9.109E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.37E-09 | 4.055E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.568E-09 | 6.069E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.498E-08 | 3.909E-05 |
|---|
| interphase | GO:0051325 |  | 7.368E-08 | 3.546E-05 |
|---|
| response to UV | GO:0009411 |  | 1.453E-06 | 5.829E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.719E-06 | 5.907E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.814E-06 | 5.455E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 1.286E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 6.181E-06 | 1.487E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 9.048E-06 | 1.979E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.786E-11 | 9.109E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.37E-09 | 4.055E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.568E-09 | 6.069E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.498E-08 | 3.909E-05 |
|---|
| interphase | GO:0051325 |  | 7.368E-08 | 3.546E-05 |
|---|
| response to UV | GO:0009411 |  | 1.453E-06 | 5.829E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.719E-06 | 5.907E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.814E-06 | 5.455E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 1.286E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 6.181E-06 | 1.487E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 9.048E-06 | 1.979E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.328E-10 | 7.655E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.232E-08 | 2.567E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.878E-08 | 3.74E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.773E-07 | 2.169E-04 |
|---|
| interphase | GO:0051325 |  | 4.14E-07 | 1.904E-04 |
|---|
| response to UV | GO:0009411 |  | 4.787E-06 | 1.835E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.45E-06 | 1.791E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.553E-06 | 1.884E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 2.734E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.426E-05 | 3.279E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.797E-05 | 5.848E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.328E-10 | 7.655E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.232E-08 | 2.567E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.878E-08 | 3.74E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.773E-07 | 2.169E-04 |
|---|
| interphase | GO:0051325 |  | 4.14E-07 | 1.904E-04 |
|---|
| response to UV | GO:0009411 |  | 4.787E-06 | 1.835E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.45E-06 | 1.791E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.553E-06 | 1.884E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 2.734E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.426E-05 | 3.279E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.797E-05 | 5.848E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.328E-10 | 7.655E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.232E-08 | 2.567E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.878E-08 | 3.74E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.773E-07 | 2.169E-04 |
|---|
| interphase | GO:0051325 |  | 4.14E-07 | 1.904E-04 |
|---|
| response to UV | GO:0009411 |  | 4.787E-06 | 1.835E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.45E-06 | 1.791E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.553E-06 | 1.884E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 2.734E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.426E-05 | 3.279E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.797E-05 | 5.848E-03 |
|---|



