Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3159-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1739 | 9.781e-03 | 1.920e-02 | 3.991e-02 |
|---|
| IPC-NIBC-129 | 0.1265 | 7.075e-02 | 4.088e-02 | 2.920e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2443 | 1.302e-01 | 1.132e-01 | 7.314e-01 |
|---|
| Loi_GPL570 | 0.2451 | 9.908e-02 | 7.408e-02 | 3.890e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1918 | 1.232e-02 | 1.680e-04 | 1.330e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2874 | 1.576e-01 | 8.642e-02 | 6.334e-01 |
|---|
| Schmidt | 0.1998 | 1.460e-02 | 2.040e-02 | 8.193e-02 |
|---|
| Sotiriou | 0.3025 | 6.679e-02 | 4.818e-02 | 4.243e-01 |
|---|
| Zhang | 0.2105 | 1.311e-02 | 1.945e-02 | 5.110e-02 |
|---|
Expression data for subnetwork 3159-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
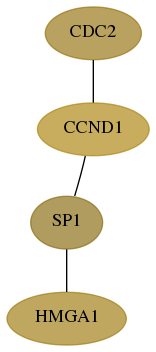
Score for each gene in subnetwork 3159-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| sp1 |   | 14 | 4 | 11 | 6 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.091 |
|---|
| hmga1 |   | 1 | 57 | 157 | 161 | 0.092 | 0.071 | 0.202 | 0.017 | 0.101 | 0.159 | 0.151 | 0.107 | -0.051 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 3159-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.158E-07 | 4.963E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.324E-06 | 1.523E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.53E-06 | 2.707E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 3.53E-06 | 2.03E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.273E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.528E-06 | 2.119E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 6.586E-06 | 2.164E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.293E-05 | 3.716E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 5.461E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.465E-05 | 5.67E-03 |
|---|
| DNA integration | GO:0015074 |  | 2.465E-05 | 5.155E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.273E-08 | 7.996E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.93E-07 | 2.358E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.697E-07 | 4.639E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.544E-07 | 5.218E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 9.133E-07 | 4.462E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.196E-06 | 4.87E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.595E-06 | 5.565E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.595E-06 | 4.869E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 2.05E-06 | 5.564E-04 |
|---|
| lysogeny | GO:0030069 |  | 3.131E-06 | 7.649E-04 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 7.736E-06 | 1.718E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 5.378E-08 | 1.294E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.17E-07 | 3.814E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.935E-07 | 6.363E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.19E-06 | 7.158E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.388E-06 | 6.679E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.666E-06 | 6.68E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.221E-06 | 7.633E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.221E-06 | 6.679E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 2.855E-06 | 7.631E-04 |
|---|
| lysogeny | GO:0030069 |  | 4.36E-06 | 1.049E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.077E-05 | 2.356E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.273E-08 | 7.996E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.93E-07 | 2.358E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.697E-07 | 4.639E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.544E-07 | 5.218E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 9.133E-07 | 4.462E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.196E-06 | 4.87E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.595E-06 | 5.565E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.595E-06 | 4.869E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 2.05E-06 | 5.564E-04 |
|---|
| lysogeny | GO:0030069 |  | 3.131E-06 | 7.649E-04 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 7.736E-06 | 1.718E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 5.378E-08 | 1.294E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.17E-07 | 3.814E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.935E-07 | 6.363E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.19E-06 | 7.158E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.388E-06 | 6.679E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.666E-06 | 6.68E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.221E-06 | 7.633E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.221E-06 | 6.679E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 2.855E-06 | 7.631E-04 |
|---|
| lysogeny | GO:0030069 |  | 4.36E-06 | 1.049E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.077E-05 | 2.356E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 5.378E-08 | 1.294E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.17E-07 | 3.814E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.935E-07 | 6.363E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.19E-06 | 7.158E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.388E-06 | 6.679E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.666E-06 | 6.68E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.221E-06 | 7.633E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.221E-06 | 6.679E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 2.855E-06 | 7.631E-04 |
|---|
| lysogeny | GO:0030069 |  | 4.36E-06 | 1.049E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.077E-05 | 2.356E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.158E-07 | 4.963E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.324E-06 | 1.523E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.53E-06 | 2.707E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 3.53E-06 | 2.03E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.273E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.528E-06 | 2.119E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 6.586E-06 | 2.164E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.293E-05 | 3.716E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 5.461E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.465E-05 | 5.67E-03 |
|---|
| DNA integration | GO:0015074 |  | 2.465E-05 | 5.155E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.158E-07 | 4.963E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.324E-06 | 1.523E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.53E-06 | 2.707E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 3.53E-06 | 2.03E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.273E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.528E-06 | 2.119E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 6.586E-06 | 2.164E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.293E-05 | 3.716E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 5.461E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.465E-05 | 5.67E-03 |
|---|
| DNA integration | GO:0015074 |  | 2.465E-05 | 5.155E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.158E-07 | 4.963E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.324E-06 | 1.523E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.53E-06 | 2.707E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 3.53E-06 | 2.03E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.273E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.528E-06 | 2.119E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 6.586E-06 | 2.164E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.293E-05 | 3.716E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 5.461E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.465E-05 | 5.67E-03 |
|---|
| DNA integration | GO:0015074 |  | 2.465E-05 | 5.155E-03 |
|---|



