Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3091-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1969 | 9.830e-03 | 1.929e-02 | 4.009e-02 |
|---|
| IPC-NIBC-129 | 0.2546 | 6.266e-02 | 3.534e-02 | 2.577e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2440 | 1.018e-02 | 5.436e-03 | 1.006e-01 |
|---|
| Loi_GPL570 | 0.2548 | 1.028e-01 | 7.777e-02 | 3.994e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2027 | 5.358e-03 | 1.400e-05 | 4.353e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3121 | 8.857e-02 | 3.915e-02 | 4.431e-01 |
|---|
| Schmidt | 0.1972 | 1.244e-02 | 1.747e-02 | 6.947e-02 |
|---|
| Sotiriou | 0.3125 | 5.551e-02 | 3.971e-02 | 3.694e-01 |
|---|
| Zhang | 0.2642 | 8.700e-03 | 1.293e-02 | 3.607e-02 |
|---|
Expression data for subnetwork 3091-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
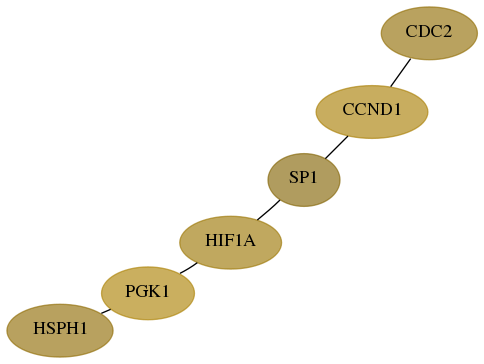
Score for each gene in subnetwork 3091-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| sp1 |   | 14 | 4 | 11 | 6 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.091 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| hsph1 |   | 2 | 35 | 17 | 17 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.161 | 0.143 | 0.132 | 0.094 |
|---|
| pgk1 |   | 4 | 19 | 1 | 4 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.310 | 0.201 | 0.127 | 0.054 |
|---|
| hif1a |   | 2 | 35 | 22 | 22 | 0.097 | 0.246 | 0.089 | -0.009 | 0.101 | 0.011 | -0.043 | 0.119 | 0.129 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 3091-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.21E-09 | 7.382E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.454E-08 | 2.822E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.691E-08 | 2.063E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.694E-08 | 3.274E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.139E-07 | 5.24E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.737E-07 | 6.659E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.817E-07 | 9.256E-05 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 5.172E-07 | 1.487E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 1.144E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 5.098E-06 | 1.173E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 5.098E-06 | 1.066E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.077E-10 | 1.485E-06 |
|---|
| glycolysis | GO:0006096 |  | 3.315E-09 | 4.05E-06 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.443E-09 | 6.061E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.001E-08 | 6.115E-06 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.645E-08 | 8.04E-06 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.728E-08 | 1.111E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.476E-08 | 1.562E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.04E-06 | 3.175E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.661E-06 | 4.509E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.661E-06 | 4.058E-04 |
|---|
| mRNA transcription | GO:0009299 |  | 1.661E-06 | 3.689E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.989E-10 | 2.403E-06 |
|---|
| glycolysis | GO:0006096 |  | 5.863E-09 | 7.054E-06 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.338E-08 | 1.073E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.645E-08 | 9.896E-06 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.997E-08 | 1.442E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.003E-08 | 2.006E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.257E-08 | 2.838E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.706E-06 | 5.13E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 2.313E-06 | 6.184E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.313E-06 | 5.566E-04 |
|---|
| mRNA transcription | GO:0009299 |  | 2.313E-06 | 5.06E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.077E-10 | 1.485E-06 |
|---|
| glycolysis | GO:0006096 |  | 3.315E-09 | 4.05E-06 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.443E-09 | 6.061E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.001E-08 | 6.115E-06 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.645E-08 | 8.04E-06 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.728E-08 | 1.111E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.476E-08 | 1.562E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.04E-06 | 3.175E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.661E-06 | 4.509E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.661E-06 | 4.058E-04 |
|---|
| mRNA transcription | GO:0009299 |  | 1.661E-06 | 3.689E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.989E-10 | 2.403E-06 |
|---|
| glycolysis | GO:0006096 |  | 5.863E-09 | 7.054E-06 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.338E-08 | 1.073E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.645E-08 | 9.896E-06 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.997E-08 | 1.442E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.003E-08 | 2.006E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.257E-08 | 2.838E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.706E-06 | 5.13E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 2.313E-06 | 6.184E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.313E-06 | 5.566E-04 |
|---|
| mRNA transcription | GO:0009299 |  | 2.313E-06 | 5.06E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.989E-10 | 2.403E-06 |
|---|
| glycolysis | GO:0006096 |  | 5.863E-09 | 7.054E-06 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.338E-08 | 1.073E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.645E-08 | 9.896E-06 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.997E-08 | 1.442E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.003E-08 | 2.006E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.257E-08 | 2.838E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.706E-06 | 5.13E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 2.313E-06 | 6.184E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.313E-06 | 5.566E-04 |
|---|
| mRNA transcription | GO:0009299 |  | 2.313E-06 | 5.06E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.21E-09 | 7.382E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.454E-08 | 2.822E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.691E-08 | 2.063E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.694E-08 | 3.274E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.139E-07 | 5.24E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.737E-07 | 6.659E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.817E-07 | 9.256E-05 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 5.172E-07 | 1.487E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 1.144E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 5.098E-06 | 1.173E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 5.098E-06 | 1.066E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.21E-09 | 7.382E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.454E-08 | 2.822E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.691E-08 | 2.063E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.694E-08 | 3.274E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.139E-07 | 5.24E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.737E-07 | 6.659E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.817E-07 | 9.256E-05 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 5.172E-07 | 1.487E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 1.144E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 5.098E-06 | 1.173E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 5.098E-06 | 1.066E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.21E-09 | 7.382E-06 |
|---|
| glycolysis | GO:0006096 |  | 2.454E-08 | 2.822E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.691E-08 | 2.063E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.694E-08 | 3.274E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.139E-07 | 5.24E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.737E-07 | 6.659E-05 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.817E-07 | 9.256E-05 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 5.172E-07 | 1.487E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 1.144E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 5.098E-06 | 1.173E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 5.098E-06 | 1.066E-03 |
|---|



