Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 27158-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2222 | 2.838e-02 | 4.954e-02 | 1.051e-01 |
|---|
| IPC-NIBC-129 | 0.0858 | 1.052e-01 | 6.577e-02 | 4.250e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1930 | 2.785e-01 | 2.719e-01 | 9.327e-01 |
|---|
| Loi_GPL570 | 0.2131 | 5.828e-01 | 6.569e-01 | 9.547e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1814 | 2.097e-03 | 1.000e-06 | 1.018e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3607 | 9.227e-01 | 9.180e-01 | 1.000e+00 |
|---|
| Schmidt | 0.0622 | 5.161e-02 | 6.875e-02 | 2.695e-01 |
|---|
| Sotiriou | 0.2223 | 7.946e-02 | 5.781e-02 | 4.796e-01 |
|---|
| Zhang | 0.0195 | 3.870e-03 | 5.745e-03 | 1.792e-02 |
|---|
Expression data for subnetwork 27158-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
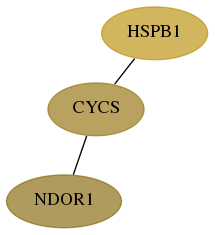
Score for each gene in subnetwork 27158-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| hspb1 |   | 5 | 13 | 118 | 99 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | 0.289 | -0.021 | 0.201 | -0.077 |
|---|
| cycs |   | 4 | 19 | 118 | 103 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | 0.272 | 0.157 | 0.077 | 0.135 |
|---|
| ndor1 |   | 1 | 57 | 190 | 190 | 0.052 | 0.053 | 0.101 | 0.023 | 0.065 | -0.010 | 0.095 | 0.139 | 0.021 |
|---|
GO Enrichment output for subnetwork 27158-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 6.772E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 2.37E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 2.572E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.929E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.895E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 3.27E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 3.402E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.853E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.524E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.463E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.211E-05 | 4.622E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.496E-10 | 3.655E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.816E-09 | 2.218E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.55E-09 | 2.891E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.111E-09 | 2.511E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 5.402E-09 | 2.639E-06 |
|---|
| nucleus organization | GO:0006997 |  | 2.634E-08 | 1.073E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 4.609E-08 | 1.609E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.223E-08 | 1.595E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.793E-08 | 2.115E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.112E-08 | 2.226E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.725E-07 | 3.83E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.459E-10 | 5.917E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.984E-09 | 3.59E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.835E-09 | 4.68E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.756E-09 | 4.064E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 8.878E-09 | 4.272E-06 |
|---|
| nucleus organization | GO:0006997 |  | 3.696E-08 | 1.482E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 7.573E-08 | 2.603E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.573E-08 | 2.278E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.148E-07 | 3.069E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.422E-07 | 3.422E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.607E-07 | 5.702E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.496E-10 | 3.655E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.816E-09 | 2.218E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.55E-09 | 2.891E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.111E-09 | 2.511E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 5.402E-09 | 2.639E-06 |
|---|
| nucleus organization | GO:0006997 |  | 2.634E-08 | 1.073E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 4.609E-08 | 1.609E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.223E-08 | 1.595E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.793E-08 | 2.115E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.112E-08 | 2.226E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.725E-07 | 3.83E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.459E-10 | 5.917E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.984E-09 | 3.59E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.835E-09 | 4.68E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.756E-09 | 4.064E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 8.878E-09 | 4.272E-06 |
|---|
| nucleus organization | GO:0006997 |  | 3.696E-08 | 1.482E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 7.573E-08 | 2.603E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.573E-08 | 2.278E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.148E-07 | 3.069E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.422E-07 | 3.422E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.607E-07 | 5.702E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.459E-10 | 5.917E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.984E-09 | 3.59E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.835E-09 | 4.68E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.756E-09 | 4.064E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 8.878E-09 | 4.272E-06 |
|---|
| nucleus organization | GO:0006997 |  | 3.696E-08 | 1.482E-05 |
|---|
| cellular component disassembly | GO:0022411 |  | 7.573E-08 | 2.603E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.573E-08 | 2.278E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.148E-07 | 3.069E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.422E-07 | 3.422E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.607E-07 | 5.702E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 6.772E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 2.37E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 2.572E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.929E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.895E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 3.27E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 3.402E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.853E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.524E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.463E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.211E-05 | 4.622E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 6.772E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 2.37E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 2.572E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.929E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.895E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 3.27E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 3.402E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.853E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.524E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.463E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.211E-05 | 4.622E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 6.772E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 2.37E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 2.572E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.929E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.895E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 3.27E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 3.402E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.853E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.524E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.463E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.211E-05 | 4.622E-03 |
|---|



