Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2596-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1721 | 1.117e-02 | 2.162e-02 | 4.516e-02 |
|---|
| IPC-NIBC-129 | 0.2033 | 7.883e-02 | 4.654e-02 | 3.251e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2379 | 2.874e-02 | 1.893e-02 | 2.679e-01 |
|---|
| Loi_GPL570 | 0.2364 | 1.363e-01 | 1.128e-01 | 4.855e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1932 | 1.315e-02 | 2.030e-04 | 1.440e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2815 | 6.430e-02 | 2.520e-02 | 3.521e-01 |
|---|
| Schmidt | 0.1765 | 1.158e-02 | 1.631e-02 | 6.448e-02 |
|---|
| Sotiriou | 0.3170 | 6.517e-02 | 4.696e-02 | 4.168e-01 |
|---|
| Zhang | 0.2935 | 1.444e-02 | 2.141e-02 | 5.545e-02 |
|---|
Expression data for subnetwork 2596-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
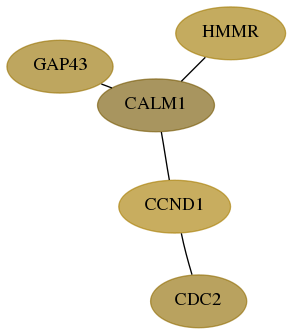
Score for each gene in subnetwork 2596-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| calm1 |   | 4 | 19 | 41 | 35 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.162 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| gap43 |   | 1 | 57 | 67 | 70 | 0.087 | 0.151 | 0.091 | -0.117 | 0.079 | 0.077 | -0.087 | 0.073 | 0.064 |
|---|
| hmmr |   | 4 | 19 | 41 | 35 | 0.112 | 0.310 | 0.147 | 0.127 | 0.145 | 0.221 | 0.179 | 0.233 | 0.144 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 2596-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| tissue regeneration | GO:0042246 |  | 4.622E-05 | 0.05315201 |
|---|
| neuron recognition | GO:0008038 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| regeneration | GO:0031099 |  | 1.01E-04 | 0.04648241 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.04752619 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.04903317 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.04678297 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.914E-04 | 0.04892289 |
|---|
| glial cell differentiation | GO:0010001 |  | 2.733E-04 | 0.06285366 |
|---|
| gliogenesis | GO:0042063 |  | 4.118E-04 | 0.08610914 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 0.01653866 |
|---|
| neuron recognition | GO:0008038 |  | 2.894E-05 | 0.03535501 |
|---|
| tissue regeneration | GO:0042246 |  | 2.894E-05 | 0.02357001 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.577E-05 | 0.02795581 |
|---|
| regeneration | GO:0031099 |  | 6.09E-05 | 0.02975349 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.815E-05 | 0.03182209 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.085E-05 | 0.03170514 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.755E-05 | 0.02978802 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.045E-04 | 0.02836103 |
|---|
| glial cell differentiation | GO:0010001 |  | 1.346E-04 | 0.03287888 |
|---|
| gliogenesis | GO:0042063 |  | 1.963E-04 | 0.04359767 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| tissue regeneration | GO:0042246 |  | 2.767E-05 | 0.0332895 |
|---|
| neuron recognition | GO:0008038 |  | 3.162E-05 | 0.02535557 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.02707358 |
|---|
| regeneration | GO:0031099 |  | 6.075E-05 | 0.02923128 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.03160619 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.03167685 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.02984009 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.141E-04 | 0.03050535 |
|---|
| glial cell differentiation | GO:0010001 |  | 1.47E-04 | 0.03536339 |
|---|
| gliogenesis | GO:0042063 |  | 2.144E-04 | 0.04688893 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 0.01653866 |
|---|
| neuron recognition | GO:0008038 |  | 2.894E-05 | 0.03535501 |
|---|
| tissue regeneration | GO:0042246 |  | 2.894E-05 | 0.02357001 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.577E-05 | 0.02795581 |
|---|
| regeneration | GO:0031099 |  | 6.09E-05 | 0.02975349 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.815E-05 | 0.03182209 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.085E-05 | 0.03170514 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.755E-05 | 0.02978802 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.045E-04 | 0.02836103 |
|---|
| glial cell differentiation | GO:0010001 |  | 1.346E-04 | 0.03287888 |
|---|
| gliogenesis | GO:0042063 |  | 1.963E-04 | 0.04359767 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| tissue regeneration | GO:0042246 |  | 2.767E-05 | 0.0332895 |
|---|
| neuron recognition | GO:0008038 |  | 3.162E-05 | 0.02535557 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.02707358 |
|---|
| regeneration | GO:0031099 |  | 6.075E-05 | 0.02923128 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.03160619 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.03167685 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.02984009 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.141E-04 | 0.03050535 |
|---|
| glial cell differentiation | GO:0010001 |  | 1.47E-04 | 0.03536339 |
|---|
| gliogenesis | GO:0042063 |  | 2.144E-04 | 0.04688893 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| tissue regeneration | GO:0042246 |  | 2.767E-05 | 0.0332895 |
|---|
| neuron recognition | GO:0008038 |  | 3.162E-05 | 0.02535557 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.02707358 |
|---|
| regeneration | GO:0031099 |  | 6.075E-05 | 0.02923128 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.03160619 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.03167685 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.02984009 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.141E-04 | 0.03050535 |
|---|
| glial cell differentiation | GO:0010001 |  | 1.47E-04 | 0.03536339 |
|---|
| gliogenesis | GO:0042063 |  | 2.144E-04 | 0.04688893 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| tissue regeneration | GO:0042246 |  | 4.622E-05 | 0.05315201 |
|---|
| neuron recognition | GO:0008038 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| regeneration | GO:0031099 |  | 1.01E-04 | 0.04648241 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.04752619 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.04903317 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.04678297 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.914E-04 | 0.04892289 |
|---|
| glial cell differentiation | GO:0010001 |  | 2.733E-04 | 0.06285366 |
|---|
| gliogenesis | GO:0042063 |  | 4.118E-04 | 0.08610914 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| tissue regeneration | GO:0042246 |  | 4.622E-05 | 0.05315201 |
|---|
| neuron recognition | GO:0008038 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| regeneration | GO:0031099 |  | 1.01E-04 | 0.04648241 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.04752619 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.04903317 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.04678297 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.914E-04 | 0.04892289 |
|---|
| glial cell differentiation | GO:0010001 |  | 2.733E-04 | 0.06285366 |
|---|
| gliogenesis | GO:0042063 |  | 4.118E-04 | 0.08610914 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| tissue regeneration | GO:0042246 |  | 4.622E-05 | 0.05315201 |
|---|
| neuron recognition | GO:0008038 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| regeneration | GO:0031099 |  | 1.01E-04 | 0.04648241 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.04752619 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.04903317 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.04678297 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.914E-04 | 0.04892289 |
|---|
| glial cell differentiation | GO:0010001 |  | 2.733E-04 | 0.06285366 |
|---|
| gliogenesis | GO:0042063 |  | 4.118E-04 | 0.08610914 |
|---|



