Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2071-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.0248 | 1.083e-02 | 2.104e-02 | 4.389e-02 |
|---|
| IPC-NIBC-129 | 0.2321 | 7.580e-02 | 4.441e-02 | 3.128e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2394 | 1.608e-02 | 9.427e-03 | 1.584e-01 |
|---|
| Loi_GPL570 | 0.2396 | 4.711e-02 | 2.717e-02 | 2.178e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1053 | 8.702e-01 | 9.991e-01 | 1.000e+00 |
|---|
| Pawitan_GPL96-GPL97 | 0.2888 | 5.612e-01 | 4.845e-01 | 9.775e-01 |
|---|
| Schmidt | 0.2531 | 1.865e-02 | 2.584e-02 | 1.049e-01 |
|---|
| Sotiriou | 0.2871 | 2.732e-01 | 2.151e-01 | 8.844e-01 |
|---|
| Zhang | 0.0831 | 1.280e-02 | 1.900e-02 | 5.010e-02 |
|---|
Expression data for subnetwork 2071-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
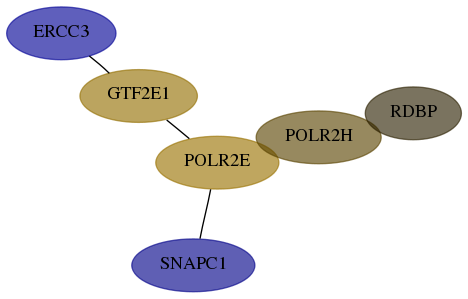
Score for each gene in subnetwork 2071-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| polr2e |   | 1 | 57 | 183 | 183 | 0.092 | 0.011 | 0.084 | 0.075 | 0.058 | 0.177 | 0.074 | 0.107 | -0.095 |
|---|
| ercc3 |   | 1 | 57 | 183 | 183 | -0.083 | 0.061 | 0.021 | 0.064 | 0.069 | 0.163 | 0.105 | 0.124 | -0.030 |
|---|
| gtf2e1 |   | 1 | 57 | 183 | 183 | 0.080 | 0.101 | 0.138 | 0.034 | 0.106 | 0.182 | 0.073 | 0.183 | 0.048 |
|---|
| polr2h |   | 1 | 57 | 183 | 183 | 0.021 | 0.282 | 0.144 | 0.302 | 0.127 | 0.129 | 0.257 | 0.153 | 0.165 |
|---|
| rdbp |   | 1 | 57 | 183 | 183 | 0.006 | 0.165 | 0.250 | 0.202 | 0.177 | 0.222 | 0.196 | 0.164 | -0.020 |
|---|
| snapc1 |   | 1 | 57 | 183 | 183 | -0.061 | 0.155 | 0.094 | 0.099 | 0.087 | 0.085 | 0.117 | 0.038 | 0.260 |
|---|
GO Enrichment output for subnetwork 2071-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.359E-12 | 5.426E-09 |
|---|
| RNA elongation | GO:0006354 |  | 2.932E-12 | 3.372E-09 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.857E-11 | 1.424E-08 |
|---|
| transcription initiation | GO:0006352 |  | 5.027E-11 | 2.89E-08 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 2.06E-06 | 9.476E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 2.06E-06 | 7.896E-04 |
|---|
| DNA topological change | GO:0006265 |  | 2.06E-06 | 6.768E-04 |
|---|
| UV protection | GO:0009650 |  | 6.175E-06 | 1.775E-03 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.864E-05 | 4.763E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 6.614E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 6.013E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.152E-13 | 2.815E-10 |
|---|
| RNA elongation | GO:0006354 |  | 1.693E-13 | 2.068E-10 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.134E-12 | 9.237E-10 |
|---|
| transcription initiation | GO:0006352 |  | 2.851E-12 | 1.741E-09 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 6.978E-07 | 3.41E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 9.303E-07 | 3.788E-04 |
|---|
| DNA topological change | GO:0006265 |  | 9.303E-07 | 3.247E-04 |
|---|
| UV protection | GO:0009650 |  | 1.827E-06 | 5.579E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 6.306E-06 | 1.712E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 8.394E-06 | 2.051E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.157E-06 | 2.034E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.384E-13 | 5.735E-10 |
|---|
| RNA elongation | GO:0006354 |  | 3.529E-13 | 4.246E-10 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.139E-12 | 1.716E-09 |
|---|
| transcription initiation | GO:0006352 |  | 5.557E-12 | 3.343E-09 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 9.719E-07 | 4.677E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 1.296E-06 | 5.196E-04 |
|---|
| DNA topological change | GO:0006265 |  | 1.296E-06 | 4.454E-04 |
|---|
| UV protection | GO:0009650 |  | 2.544E-06 | 7.652E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 8.781E-06 | 2.347E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.169E-05 | 2.812E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.275E-05 | 2.789E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.152E-13 | 2.815E-10 |
|---|
| RNA elongation | GO:0006354 |  | 1.693E-13 | 2.068E-10 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.134E-12 | 9.237E-10 |
|---|
| transcription initiation | GO:0006352 |  | 2.851E-12 | 1.741E-09 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 6.978E-07 | 3.41E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 9.303E-07 | 3.788E-04 |
|---|
| DNA topological change | GO:0006265 |  | 9.303E-07 | 3.247E-04 |
|---|
| UV protection | GO:0009650 |  | 1.827E-06 | 5.579E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 6.306E-06 | 1.712E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 8.394E-06 | 2.051E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.157E-06 | 2.034E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.384E-13 | 5.735E-10 |
|---|
| RNA elongation | GO:0006354 |  | 3.529E-13 | 4.246E-10 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.139E-12 | 1.716E-09 |
|---|
| transcription initiation | GO:0006352 |  | 5.557E-12 | 3.343E-09 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 9.719E-07 | 4.677E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 1.296E-06 | 5.196E-04 |
|---|
| DNA topological change | GO:0006265 |  | 1.296E-06 | 4.454E-04 |
|---|
| UV protection | GO:0009650 |  | 2.544E-06 | 7.652E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 8.781E-06 | 2.347E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.169E-05 | 2.812E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.275E-05 | 2.789E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.384E-13 | 5.735E-10 |
|---|
| RNA elongation | GO:0006354 |  | 3.529E-13 | 4.246E-10 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.139E-12 | 1.716E-09 |
|---|
| transcription initiation | GO:0006352 |  | 5.557E-12 | 3.343E-09 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 9.719E-07 | 4.677E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 1.296E-06 | 5.196E-04 |
|---|
| DNA topological change | GO:0006265 |  | 1.296E-06 | 4.454E-04 |
|---|
| UV protection | GO:0009650 |  | 2.544E-06 | 7.652E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 8.781E-06 | 2.347E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.169E-05 | 2.812E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.275E-05 | 2.789E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.359E-12 | 5.426E-09 |
|---|
| RNA elongation | GO:0006354 |  | 2.932E-12 | 3.372E-09 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.857E-11 | 1.424E-08 |
|---|
| transcription initiation | GO:0006352 |  | 5.027E-11 | 2.89E-08 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 2.06E-06 | 9.476E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 2.06E-06 | 7.896E-04 |
|---|
| DNA topological change | GO:0006265 |  | 2.06E-06 | 6.768E-04 |
|---|
| UV protection | GO:0009650 |  | 6.175E-06 | 1.775E-03 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.864E-05 | 4.763E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 6.614E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 6.013E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.359E-12 | 5.426E-09 |
|---|
| RNA elongation | GO:0006354 |  | 2.932E-12 | 3.372E-09 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.857E-11 | 1.424E-08 |
|---|
| transcription initiation | GO:0006352 |  | 5.027E-11 | 2.89E-08 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 2.06E-06 | 9.476E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 2.06E-06 | 7.896E-04 |
|---|
| DNA topological change | GO:0006265 |  | 2.06E-06 | 6.768E-04 |
|---|
| UV protection | GO:0009650 |  | 6.175E-06 | 1.775E-03 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.864E-05 | 4.763E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 6.614E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.359E-12 | 5.426E-09 |
|---|
| RNA elongation | GO:0006354 |  | 2.932E-12 | 3.372E-09 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.857E-11 | 1.424E-08 |
|---|
| transcription initiation | GO:0006352 |  | 5.027E-11 | 2.89E-08 |
|---|
| nucleotide-excision repair. DNA incision | GO:0033683 |  | 2.06E-06 | 9.476E-04 |
|---|
| transcription-coupled nucleotide-excision repair | GO:0006283 |  | 2.06E-06 | 7.896E-04 |
|---|
| DNA topological change | GO:0006265 |  | 2.06E-06 | 6.768E-04 |
|---|
| UV protection | GO:0009650 |  | 6.175E-06 | 1.775E-03 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.864E-05 | 4.763E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.876E-05 | 6.614E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 6.013E-03 |
|---|



