Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1871-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1143 | 1.141e-02 | 2.203e-02 | 4.604e-02 |
|---|
| IPC-NIBC-129 | 0.2348 | 6.039e-02 | 3.382e-02 | 2.479e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2369 | 1.521e-02 | 8.819e-03 | 1.502e-01 |
|---|
| Loi_GPL570 | 0.2578 | 5.659e-02 | 3.487e-02 | 2.531e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1707 | 9.412e-02 | 4.033e-02 | 7.699e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3058 | 1.539e-01 | 8.362e-02 | 6.251e-01 |
|---|
| Schmidt | 0.2401 | 1.038e-02 | 1.466e-02 | 5.744e-02 |
|---|
| Sotiriou | 0.3239 | 9.508e-02 | 6.981e-02 | 5.398e-01 |
|---|
| Zhang | 0.2127 | 9.661e-03 | 1.436e-02 | 3.944e-02 |
|---|
Expression data for subnetwork 1871-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
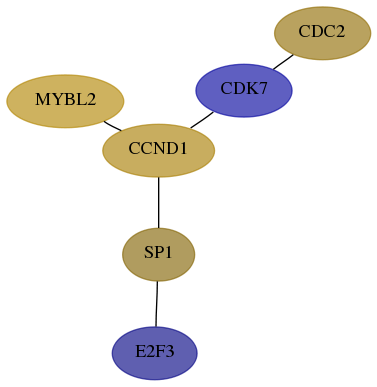
Score for each gene in subnetwork 1871-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| sp1 |   | 14 | 4 | 11 | 6 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.091 |
|---|
| e2f3 |   | 1 | 57 | 81 | 85 | -0.054 | 0.341 | 0.099 | 0.140 | 0.072 | 0.097 | 0.340 | 0.081 | 0.032 |
|---|
| mybl2 |   | 9 | 7 | 29 | 13 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.275 | 0.202 | 0.182 | 0.074 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| cdk7 |   | 2 | 35 | 81 | 76 | -0.102 | 0.121 | 0.116 | 0.126 | 0.170 | 0.090 | -0.085 | 0.182 | 0.021 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 1871-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 4.366E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 9.914E-06 | 5.7E-03 |
|---|
| transcription initiation | GO:0006352 |  | 1.78E-05 | 8.186E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 0.01442324 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.01412404 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 5.477E-05 | 0.01574632 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.51E-05 | 0.01919142 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.01727228 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.01890427 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.785E-07 | 9.246E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.393E-07 | 7.809E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.703E-07 | 7.087E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.305E-06 | 7.971E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.857E-06 | 9.071E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 9.917E-04 |
|---|
| transcription initiation | GO:0006352 |  | 3.2E-06 | 1.117E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.181E-05 | 3.607E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.649E-05 | 4.477E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.005E-05 | 4.897E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.195E-05 | 4.875E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.213E-07 | 1.495E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.049E-06 | 1.262E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.212E-06 | 9.721E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.818E-06 | 1.093E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.716E-06 | 1.307E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.392E-06 | 1.36E-03 |
|---|
| transcription initiation | GO:0006352 |  | 4.771E-06 | 1.64E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.644E-05 | 4.946E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.067E-05 | 5.525E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.79E-05 | 6.714E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.056E-05 | 6.683E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.785E-07 | 9.246E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.393E-07 | 7.809E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.703E-07 | 7.087E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.305E-06 | 7.971E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.857E-06 | 9.071E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 9.917E-04 |
|---|
| transcription initiation | GO:0006352 |  | 3.2E-06 | 1.117E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.181E-05 | 3.607E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.649E-05 | 4.477E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.005E-05 | 4.897E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.195E-05 | 4.875E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.213E-07 | 1.495E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.049E-06 | 1.262E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.212E-06 | 9.721E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.818E-06 | 1.093E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.716E-06 | 1.307E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.392E-06 | 1.36E-03 |
|---|
| transcription initiation | GO:0006352 |  | 4.771E-06 | 1.64E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.644E-05 | 4.946E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.067E-05 | 5.525E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.79E-05 | 6.714E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.056E-05 | 6.683E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.213E-07 | 1.495E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.049E-06 | 1.262E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.212E-06 | 9.721E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.818E-06 | 1.093E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.716E-06 | 1.307E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.392E-06 | 1.36E-03 |
|---|
| transcription initiation | GO:0006352 |  | 4.771E-06 | 1.64E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.644E-05 | 4.946E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.067E-05 | 5.525E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.79E-05 | 6.714E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.056E-05 | 6.683E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 4.366E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 9.914E-06 | 5.7E-03 |
|---|
| transcription initiation | GO:0006352 |  | 1.78E-05 | 8.186E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 0.01442324 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.01412404 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 5.477E-05 | 0.01574632 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.51E-05 | 0.01919142 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.01727228 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.01890427 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 4.366E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 9.914E-06 | 5.7E-03 |
|---|
| transcription initiation | GO:0006352 |  | 1.78E-05 | 8.186E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 0.01442324 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.01412404 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 5.477E-05 | 0.01574632 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.51E-05 | 0.01919142 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.01727228 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.01890427 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 5.962E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 4.366E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 5.785E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 9.914E-06 | 5.7E-03 |
|---|
| transcription initiation | GO:0006352 |  | 1.78E-05 | 8.186E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.763E-05 | 0.01442324 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.299E-05 | 0.01412404 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 5.477E-05 | 0.01574632 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.51E-05 | 0.01919142 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.51E-05 | 0.01727228 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.041E-05 | 0.01890427 |
|---|



