Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1780-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2070 | 6.258e-03 | 1.288e-02 | 2.621e-02 |
|---|
| IPC-NIBC-129 | 0.2188 | 5.066e-02 | 2.739e-02 | 2.051e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2656 | 2.105e-02 | 1.303e-02 | 2.037e-01 |
|---|
| Loi_GPL570 | 0.2718 | 6.929e-02 | 4.587e-02 | 2.972e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1634 | 3.684e-03 | 5.000e-06 | 2.491e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3632 | 9.115e-02 | 4.073e-02 | 4.518e-01 |
|---|
| Schmidt | 0.2256 | 1.488e-02 | 2.078e-02 | 8.353e-02 |
|---|
| Sotiriou | 0.3013 | 1.074e-01 | 7.933e-02 | 5.819e-01 |
|---|
| Zhang | 0.2615 | 3.707e-03 | 5.501e-03 | 1.726e-02 |
|---|
Expression data for subnetwork 1780-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
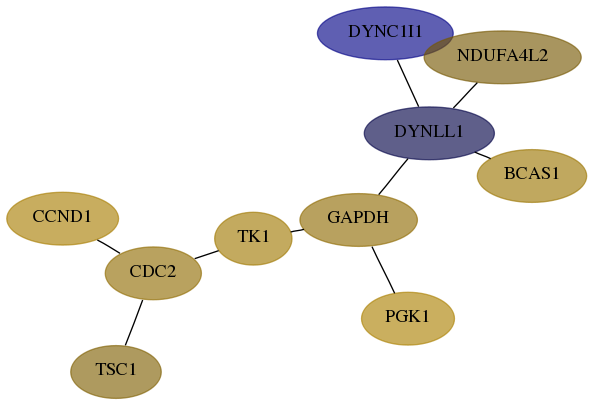
Score for each gene in subnetwork 1780-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| bcas1 |   | 1 | 57 | 1 | 10 | 0.096 | -0.141 | 0.075 | 0.173 | 0.136 | 0.053 | 0.065 | 0.068 | 0.062 |
|---|
| pgk1 |   | 4 | 19 | 1 | 4 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.310 | 0.201 | 0.127 | 0.054 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
| tsc1 |   | 11 | 6 | 1 | 3 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.196 | 0.006 | 0.010 | -0.007 |
|---|
| ndufa4l2 |   | 1 | 57 | 1 | 10 | 0.039 | 0.271 | 0.157 | 0.052 | 0.136 | 0.281 | 0.142 | 0.177 | 0.139 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| gapdh |   | 2 | 35 | 1 | 7 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.288 | 0.064 | 0.213 | -0.088 |
|---|
| tk1 |   | 4 | 19 | 1 | 4 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.257 | 0.204 | 0.142 | 0.164 |
|---|
| dync1i1 |   | 1 | 57 | 1 | 10 | -0.058 | 0.102 | 0.055 | 0.124 | 0.054 | 0.056 | 0.082 | 0.036 | 0.166 |
|---|
| dynll1 |   | 2 | 35 | 1 | 7 | -0.012 | 0.108 | 0.088 | 0.087 | 0.098 | 0.128 | 0.113 | 0.093 | 0.074 |
|---|
GO Enrichment output for subnetwork 1780-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 3.235E-12 | 7.44E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.177E-11 | 1.353E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.395E-11 | 2.603E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.459E-11 | 3.714E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 6.201E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 1.301E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 3.008E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 2.205E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.995E-06 | 2.554E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 9.995E-06 | 2.299E-03 |
|---|
| cell projection assembly | GO:0030031 |  | 2.079E-05 | 4.347E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 5.818E-13 | 1.421E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.007E-12 | 2.452E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 6.73E-12 | 5.48E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.452E-11 | 8.867E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.078E-11 | 1.504E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.071E-09 | 1.25E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.052E-08 | 1.763E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.743E-06 | 1.448E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.112E-06 | 1.93E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 3.24E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.326E-05 | 2.946E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 5.763E-13 | 1.387E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.042E-12 | 2.456E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 6.991E-12 | 5.607E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.525E-11 | 9.174E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.265E-11 | 1.571E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.38E-09 | 1.355E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.561E-08 | 1.911E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 1.53E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.36E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.422E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 3.111E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 5.818E-13 | 1.421E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.007E-12 | 2.452E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 6.73E-12 | 5.48E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.452E-11 | 8.867E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.078E-11 | 1.504E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.071E-09 | 1.25E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.052E-08 | 1.763E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.743E-06 | 1.448E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.112E-06 | 1.93E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 3.24E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.326E-05 | 2.946E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 5.763E-13 | 1.387E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.042E-12 | 2.456E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 6.991E-12 | 5.607E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.525E-11 | 9.174E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.265E-11 | 1.571E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.38E-09 | 1.355E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.561E-08 | 1.911E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 1.53E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.36E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.422E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 3.111E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 5.763E-13 | 1.387E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.042E-12 | 2.456E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 6.991E-12 | 5.607E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.525E-11 | 9.174E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.265E-11 | 1.571E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.38E-09 | 1.355E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.561E-08 | 1.911E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 1.53E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.36E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.422E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 3.111E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 3.235E-12 | 7.44E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.177E-11 | 1.353E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.395E-11 | 2.603E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.459E-11 | 3.714E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 6.201E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 1.301E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 3.008E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 2.205E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.995E-06 | 2.554E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 9.995E-06 | 2.299E-03 |
|---|
| cell projection assembly | GO:0030031 |  | 2.079E-05 | 4.347E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 3.235E-12 | 7.44E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.177E-11 | 1.353E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.395E-11 | 2.603E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.459E-11 | 3.714E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 6.201E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 1.301E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 3.008E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 2.205E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.995E-06 | 2.554E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 9.995E-06 | 2.299E-03 |
|---|
| cell projection assembly | GO:0030031 |  | 2.079E-05 | 4.347E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 3.235E-12 | 7.44E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.177E-11 | 1.353E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.395E-11 | 2.603E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.459E-11 | 3.714E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.348E-10 | 6.201E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.393E-10 | 1.301E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 9.153E-09 | 3.008E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.669E-08 | 2.205E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.995E-06 | 2.554E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 9.995E-06 | 2.299E-03 |
|---|
| cell projection assembly | GO:0030031 |  | 2.079E-05 | 4.347E-03 |
|---|



