Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1537-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2041 | 4.403e-02 | 7.306e-02 | 1.538e-01 |
|---|
| IPC-NIBC-129 | 0.0863 | 8.058e-02 | 4.778e-02 | 3.321e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1716 | 2.763e-01 | 2.695e-01 | 9.313e-01 |
|---|
| Loi_GPL570 | 0.2346 | 4.537e-01 | 5.006e-01 | 8.952e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2114 | 4.114e-03 | 6.000e-06 | 2.946e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3808 | 9.662e-01 | 9.672e-01 | 1.000e+00 |
|---|
| Schmidt | 0.0836 | 3.807e-02 | 5.136e-02 | 2.065e-01 |
|---|
| Sotiriou | 0.2418 | 4.790e-02 | 3.405e-02 | 3.289e-01 |
|---|
| Zhang | 0.0108 | 2.762e-03 | 4.092e-03 | 1.334e-02 |
|---|
Expression data for subnetwork 1537-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
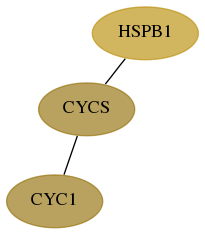
Score for each gene in subnetwork 1537-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| hspb1 |   | 5 | 13 | 118 | 99 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | 0.289 | -0.021 | 0.201 | -0.077 |
|---|
| cycs |   | 4 | 19 | 118 | 103 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | 0.272 | 0.157 | 0.077 | 0.135 |
|---|
| cyc1 |   | 1 | 57 | 189 | 189 | 0.072 | 0.077 | 0.066 | 0.193 | 0.165 | 0.253 | 0.104 | 0.189 | 0.064 |
|---|
GO Enrichment output for subnetwork 1537-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| electron transport chain | GO:0022900 |  | 1.506E-07 | 3.463E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 3.386E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 1.58E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 1.929E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.543E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.579E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 2.803E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 2.977E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.313E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.071E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.057E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 7.481E-11 | 1.828E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 9.081E-10 | 1.109E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.776E-09 | 1.446E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.056E-09 | 1.256E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.702E-09 | 1.32E-06 |
|---|
| nucleus organization | GO:0006997 |  | 1.318E-08 | 5.367E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.307E-08 | 8.051E-06 |
|---|
| activation of caspase activity | GO:0006919 |  | 2.614E-08 | 7.982E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.901E-08 | 1.059E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 4.562E-08 | 1.114E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 8.636E-08 | 1.918E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.23E-10 | 2.959E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.493E-09 | 1.796E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.919E-09 | 2.341E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.38E-09 | 2.033E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.441E-09 | 2.137E-06 |
|---|
| nucleus organization | GO:0006997 |  | 1.85E-08 | 7.418E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.791E-08 | 1.303E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.791E-08 | 1.14E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.748E-08 | 1.537E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 7.121E-08 | 1.713E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.306E-07 | 2.856E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 7.481E-11 | 1.828E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 9.081E-10 | 1.109E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.776E-09 | 1.446E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.056E-09 | 1.256E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.702E-09 | 1.32E-06 |
|---|
| nucleus organization | GO:0006997 |  | 1.318E-08 | 5.367E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.307E-08 | 8.051E-06 |
|---|
| activation of caspase activity | GO:0006919 |  | 2.614E-08 | 7.982E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.901E-08 | 1.059E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 4.562E-08 | 1.114E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 8.636E-08 | 1.918E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.23E-10 | 2.959E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.493E-09 | 1.796E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.919E-09 | 2.341E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.38E-09 | 2.033E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.441E-09 | 2.137E-06 |
|---|
| nucleus organization | GO:0006997 |  | 1.85E-08 | 7.418E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.791E-08 | 1.303E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.791E-08 | 1.14E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.748E-08 | 1.537E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 7.121E-08 | 1.713E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.306E-07 | 2.856E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.23E-10 | 2.959E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.493E-09 | 1.796E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.919E-09 | 2.341E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.38E-09 | 2.033E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.441E-09 | 2.137E-06 |
|---|
| nucleus organization | GO:0006997 |  | 1.85E-08 | 7.418E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.791E-08 | 1.303E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.791E-08 | 1.14E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.748E-08 | 1.537E-05 |
|---|
| DNA catabolic process | GO:0006308 |  | 7.121E-08 | 1.713E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.306E-07 | 2.856E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| electron transport chain | GO:0022900 |  | 1.506E-07 | 3.463E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 3.386E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 1.58E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 1.929E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.543E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.579E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 2.803E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 2.977E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.313E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.071E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.057E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| electron transport chain | GO:0022900 |  | 1.506E-07 | 3.463E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 3.386E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 1.58E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 1.929E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.543E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.579E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 2.803E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 2.977E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.313E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.071E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.057E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| electron transport chain | GO:0022900 |  | 1.506E-07 | 3.463E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.945E-07 | 3.386E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.06E-06 | 1.58E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.355E-06 | 1.929E-03 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.355E-06 | 1.543E-03 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.12E-06 | 1.579E-03 |
|---|
| response to heat | GO:0009408 |  | 8.531E-06 | 2.803E-03 |
|---|
| nucleus organization | GO:0006997 |  | 1.035E-05 | 2.977E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.688E-05 | 4.313E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.77E-05 | 4.071E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.941E-05 | 4.057E-03 |
|---|



