Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1459-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1768 | 1.262e-02 | 2.410e-02 | 5.054e-02 |
|---|
| IPC-NIBC-129 | 0.1688 | 1.464e-01 | 9.789e-02 | 5.571e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2321 | 5.707e-02 | 4.286e-02 | 4.554e-01 |
|---|
| Loi_GPL570 | 0.1860 | 1.099e-01 | 8.500e-02 | 4.189e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2128 | 1.110e-02 | 1.240e-04 | 1.169e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3144 | 1.699e-01 | 9.579e-02 | 6.595e-01 |
|---|
| Schmidt | 0.1923 | 1.661e-02 | 2.311e-02 | 9.339e-02 |
|---|
| Sotiriou | 0.2944 | 4.675e-02 | 3.320e-02 | 3.225e-01 |
|---|
| Zhang | 0.2033 | 8.377e-03 | 1.245e-02 | 3.492e-02 |
|---|
Expression data for subnetwork 1459-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
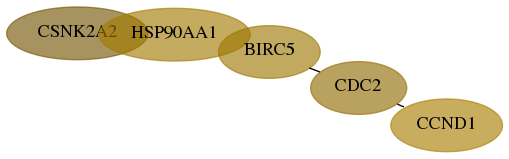
Score for each gene in subnetwork 1459-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| csnk2a2 |   | 1 | 57 | 158 | 162 | 0.036 | 0.182 | 0.089 | -0.033 | 0.074 | 0.149 | 0.060 | 0.090 | -0.155 |
|---|
| birc5 |   | 8 | 11 | 37 | 28 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.202 | 0.236 | 0.241 | 0.205 |
|---|
| hsp90aa1 |   | 6 | 12 | 37 | 29 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.169 | 0.069 | 0.102 | -0.011 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 1459-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.469E-06 | 7.439E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.469E-06 | 4.96E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.469E-06 | 3.72E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 4.165E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 9.053E-06 | 3.47E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 9.053E-06 | 2.975E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.207E-05 | 3.469E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.551E-05 | 3.963E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.689E-05 | 3.884E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.046E-05 | 4.277E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.041E-07 | 1.232E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.566E-06 | 1.913E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.192E-06 | 1.785E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.192E-06 | 1.339E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 1.428E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.922E-06 | 1.19E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.922E-06 | 1.02E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.922E-06 | 8.924E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 3.397E-06 | 9.221E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 4.127E-06 | 1.008E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.391E-06 | 9.753E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 1.991E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.181E-06 | 2.624E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.053E-06 | 2.448E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.053E-06 | 1.836E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.958E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.07E-06 | 1.632E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.07E-06 | 1.399E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.07E-06 | 1.224E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.686E-06 | 1.253E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.756E-06 | 1.385E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.552E-06 | 1.433E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.041E-07 | 1.232E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.566E-06 | 1.913E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.192E-06 | 1.785E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.192E-06 | 1.339E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 1.428E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.922E-06 | 1.19E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.922E-06 | 1.02E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.922E-06 | 8.924E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 3.397E-06 | 9.221E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 4.127E-06 | 1.008E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.391E-06 | 9.753E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 1.991E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.181E-06 | 2.624E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.053E-06 | 2.448E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.053E-06 | 1.836E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.958E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.07E-06 | 1.632E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.07E-06 | 1.399E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.07E-06 | 1.224E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.686E-06 | 1.253E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.756E-06 | 1.385E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.552E-06 | 1.433E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 1.991E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.181E-06 | 2.624E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.053E-06 | 2.448E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.053E-06 | 1.836E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.958E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.07E-06 | 1.632E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.07E-06 | 1.399E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.07E-06 | 1.224E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.686E-06 | 1.253E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.756E-06 | 1.385E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.552E-06 | 1.433E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.469E-06 | 7.439E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.469E-06 | 4.96E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.469E-06 | 3.72E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 4.165E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 9.053E-06 | 3.47E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 9.053E-06 | 2.975E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.207E-05 | 3.469E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.551E-05 | 3.963E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.689E-05 | 3.884E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.046E-05 | 4.277E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.469E-06 | 7.439E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.469E-06 | 4.96E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.469E-06 | 3.72E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 4.165E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 9.053E-06 | 3.47E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 9.053E-06 | 2.975E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.207E-05 | 3.469E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.551E-05 | 3.963E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.689E-05 | 3.884E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.046E-05 | 4.277E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.469E-06 | 7.439E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.469E-06 | 4.96E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 6.469E-06 | 3.72E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 4.165E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 9.053E-06 | 3.47E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 9.053E-06 | 2.975E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.207E-05 | 3.469E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.551E-05 | 3.963E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.689E-05 | 3.884E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.046E-05 | 4.277E-03 |
|---|



