Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1164-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1368 | 1.654e-02 | 3.067e-02 | 6.476e-02 |
|---|
| IPC-NIBC-129 | 0.1618 | 1.095e-01 | 6.904e-02 | 4.402e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2191 | 6.553e-02 | 5.049e-02 | 4.998e-01 |
|---|
| Loi_GPL570 | 0.2098 | 1.141e-01 | 8.929e-02 | 4.300e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2525 | 4.492e-02 | 6.227e-03 | 4.913e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2526 | 4.036e-02 | 1.326e-02 | 2.439e-01 |
|---|
| Schmidt | 0.1896 | 2.565e-02 | 3.514e-02 | 1.431e-01 |
|---|
| Sotiriou | 0.2670 | 2.363e-02 | 1.635e-02 | 1.774e-01 |
|---|
| Zhang | 0.3356 | 2.327e-02 | 3.432e-02 | 8.260e-02 |
|---|
Expression data for subnetwork 1164-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
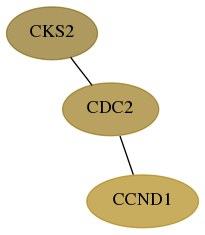
Score for each gene in subnetwork 1164-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cks2 |   | 1 | 57 | 134 | 136 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.167 | 0.263 | 0.151 | 0.284 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 1164-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.649E-07 | 1.069E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.06E-06 | 2.369E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.175E-05 | 9.012E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.056E-05 | 0.0118193 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.476E-05 | 0.01138808 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.7E-05 | 0.01035122 |
|---|
| spindle organization | GO:0007051 |  | 4.252E-05 | 0.01397099 |
|---|
| meiosis I | GO:0007127 |  | 4.847E-05 | 0.01393461 |
|---|
| response to calcium ion | GO:0051592 |  | 7.613E-05 | 0.01945471 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.809E-05 | 0.02026103 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.655E-05 | 0.02018755 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.793E-08 | 1.904E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.646E-07 | 8.118E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.506E-06 | 3.669E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.704E-06 | 4.705E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.959E-06 | 4.377E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.622E-06 | 3.918E-03 |
|---|
| spindle organization | GO:0007051 |  | 1.175E-05 | 4.101E-03 |
|---|
| meiosis I | GO:0007127 |  | 1.665E-05 | 5.084E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.45E-05 | 6.649E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.669E-05 | 6.521E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 2.783E-05 | 6.18E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.28E-07 | 3.08E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.256E-07 | 1.114E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.647E-06 | 4.529E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.903E-06 | 5.956E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.158E-05 | 5.574E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.247E-05 | 5.002E-03 |
|---|
| spindle organization | GO:0007051 |  | 1.636E-05 | 5.624E-03 |
|---|
| meiosis I | GO:0007127 |  | 2.196E-05 | 6.605E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.41E-05 | 9.116E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 3.561E-05 | 8.568E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.716E-05 | 8.127E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.793E-08 | 1.904E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.646E-07 | 8.118E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.506E-06 | 3.669E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.704E-06 | 4.705E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.959E-06 | 4.377E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.622E-06 | 3.918E-03 |
|---|
| spindle organization | GO:0007051 |  | 1.175E-05 | 4.101E-03 |
|---|
| meiosis I | GO:0007127 |  | 1.665E-05 | 5.084E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.45E-05 | 6.649E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.669E-05 | 6.521E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 2.783E-05 | 6.18E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.28E-07 | 3.08E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.256E-07 | 1.114E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.647E-06 | 4.529E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.903E-06 | 5.956E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.158E-05 | 5.574E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.247E-05 | 5.002E-03 |
|---|
| spindle organization | GO:0007051 |  | 1.636E-05 | 5.624E-03 |
|---|
| meiosis I | GO:0007127 |  | 2.196E-05 | 6.605E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.41E-05 | 9.116E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 3.561E-05 | 8.568E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.716E-05 | 8.127E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.28E-07 | 3.08E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.256E-07 | 1.114E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.647E-06 | 4.529E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.903E-06 | 5.956E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.158E-05 | 5.574E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.247E-05 | 5.002E-03 |
|---|
| spindle organization | GO:0007051 |  | 1.636E-05 | 5.624E-03 |
|---|
| meiosis I | GO:0007127 |  | 2.196E-05 | 6.605E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.41E-05 | 9.116E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 3.561E-05 | 8.568E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.716E-05 | 8.127E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.649E-07 | 1.069E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.06E-06 | 2.369E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.175E-05 | 9.012E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.056E-05 | 0.0118193 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.476E-05 | 0.01138808 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.7E-05 | 0.01035122 |
|---|
| spindle organization | GO:0007051 |  | 4.252E-05 | 0.01397099 |
|---|
| meiosis I | GO:0007127 |  | 4.847E-05 | 0.01393461 |
|---|
| response to calcium ion | GO:0051592 |  | 7.613E-05 | 0.01945471 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.809E-05 | 0.02026103 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.655E-05 | 0.02018755 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.649E-07 | 1.069E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.06E-06 | 2.369E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.175E-05 | 9.012E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.056E-05 | 0.0118193 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.476E-05 | 0.01138808 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.7E-05 | 0.01035122 |
|---|
| spindle organization | GO:0007051 |  | 4.252E-05 | 0.01397099 |
|---|
| meiosis I | GO:0007127 |  | 4.847E-05 | 0.01393461 |
|---|
| response to calcium ion | GO:0051592 |  | 7.613E-05 | 0.01945471 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.809E-05 | 0.02026103 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.655E-05 | 0.02018755 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.649E-07 | 1.069E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.06E-06 | 2.369E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.175E-05 | 9.012E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.056E-05 | 0.0118193 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.476E-05 | 0.01138808 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.7E-05 | 0.01035122 |
|---|
| spindle organization | GO:0007051 |  | 4.252E-05 | 0.01397099 |
|---|
| meiosis I | GO:0007127 |  | 4.847E-05 | 0.01393461 |
|---|
| response to calcium ion | GO:0051592 |  | 7.613E-05 | 0.01945471 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.809E-05 | 0.02026103 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.655E-05 | 0.02018755 |
|---|



