Study run-b2
Study informations
100 subnetworks in total page | file
190 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1022-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1248 | 9.509e-03 | 1.872e-02 | 3.887e-02 |
|---|
| IPC-NIBC-129 | 0.1806 | 9.224e-02 | 5.620e-02 | 3.775e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2456 | 4.520e-02 | 3.249e-02 | 3.850e-01 |
|---|
| Loi_GPL570 | 0.2237 | 2.498e-01 | 2.452e-01 | 6.967e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2466 | 6.684e-02 | 1.740e-02 | 6.444e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3352 | 7.037e-02 | 2.853e-02 | 3.764e-01 |
|---|
| Schmidt | 0.1310 | 1.498e-02 | 2.091e-02 | 8.410e-02 |
|---|
| Sotiriou | 0.3009 | 2.616e-02 | 1.816e-02 | 1.950e-01 |
|---|
| Zhang | 0.2853 | 5.918e-03 | 8.798e-03 | 2.590e-02 |
|---|
Expression data for subnetwork 1022-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Zhang |
Subnetwork structure for each dataset
- Desmedt
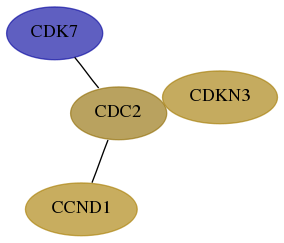
Score for each gene in subnetwork 1022-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Zhang |
|---|
| cdkn3 |   | 12 | 5 | 31 | 16 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.323 | 0.186 | 0.198 | 0.271 |
|---|
| cdc2 |   | 83 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.297 |
|---|
| cdk7 |   | 2 | 35 | 81 | 76 | -0.102 | 0.121 | 0.116 | 0.126 | 0.170 | 0.090 | -0.085 | 0.182 | 0.021 |
|---|
| ccnd1 |   | 76 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.144 |
|---|
GO Enrichment output for subnetwork 1022-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.234E-09 | 5.138E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.818E-07 | 5.541E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.883E-06 | 2.211E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.966E-06 | 2.281E-03 |
|---|
| interphase | GO:0051325 |  | 4.251E-06 | 1.956E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.645E-05 | 6.305E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 9.449E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.876E-05 | 8.268E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.463E-05 | 8.85E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.777E-05 | 8.688E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.214E-05 | 0.01508317 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.139E-10 | 1.011E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.131E-07 | 1.381E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.127E-06 | 9.176E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.24E-06 | 7.575E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 8.405E-06 | 4.107E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.119E-05 | 4.555E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.437E-05 | 5.014E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.671E-05 | 5.102E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.794E-05 | 4.87E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.941E-05 | 7.184E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.974E-05 | 0.01104737 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.017E-10 | 9.665E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.162E-07 | 1.398E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.054E-06 | 8.457E-04 |
|---|
| interphase | GO:0051325 |  | 1.158E-06 | 6.967E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.296E-06 | 6.235E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.903E-06 | 3.169E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.169E-05 | 4.017E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.386E-05 | 4.167E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.621E-05 | 4.333E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.745E-05 | 4.199E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.906E-05 | 6.357E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.139E-10 | 1.011E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.131E-07 | 1.381E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.127E-06 | 9.176E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.24E-06 | 7.575E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 8.405E-06 | 4.107E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.119E-05 | 4.555E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.437E-05 | 5.014E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.671E-05 | 5.102E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.794E-05 | 4.87E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.941E-05 | 7.184E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.974E-05 | 0.01104737 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.017E-10 | 9.665E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.162E-07 | 1.398E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.054E-06 | 8.457E-04 |
|---|
| interphase | GO:0051325 |  | 1.158E-06 | 6.967E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.296E-06 | 6.235E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.903E-06 | 3.169E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.169E-05 | 4.017E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.386E-05 | 4.167E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.621E-05 | 4.333E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.745E-05 | 4.199E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.906E-05 | 6.357E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.017E-10 | 9.665E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.162E-07 | 1.398E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.054E-06 | 8.457E-04 |
|---|
| interphase | GO:0051325 |  | 1.158E-06 | 6.967E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.296E-06 | 6.235E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.903E-06 | 3.169E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.169E-05 | 4.017E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.386E-05 | 4.167E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.621E-05 | 4.333E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.745E-05 | 4.199E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.906E-05 | 6.357E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.234E-09 | 5.138E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.818E-07 | 5.541E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.883E-06 | 2.211E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.966E-06 | 2.281E-03 |
|---|
| interphase | GO:0051325 |  | 4.251E-06 | 1.956E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.645E-05 | 6.305E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 9.449E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.876E-05 | 8.268E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.463E-05 | 8.85E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.777E-05 | 8.688E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.214E-05 | 0.01508317 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.234E-09 | 5.138E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.818E-07 | 5.541E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.883E-06 | 2.211E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.966E-06 | 2.281E-03 |
|---|
| interphase | GO:0051325 |  | 4.251E-06 | 1.956E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.645E-05 | 6.305E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 9.449E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.876E-05 | 8.268E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.463E-05 | 8.85E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.777E-05 | 8.688E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.214E-05 | 0.01508317 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.234E-09 | 5.138E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.818E-07 | 5.541E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.883E-06 | 2.211E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.966E-06 | 2.281E-03 |
|---|
| interphase | GO:0051325 |  | 4.251E-06 | 1.956E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.645E-05 | 6.305E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 2.876E-05 | 9.449E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.876E-05 | 8.268E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.463E-05 | 8.85E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.777E-05 | 8.688E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.214E-05 | 0.01508317 |
|---|



