Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 8721-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1635 | 1.541e-02 | 5.833e-03 | 8.199e-02 |
|---|
| IPC-NIBC-129 | 0.2013 | 6.008e-02 | 4.934e-02 | 2.829e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2261 | 3.126e-02 | 2.316e-02 | 3.158e-01 |
|---|
| Loi_GPL570 | 0.2585 | 1.259e-01 | 1.548e-01 | 4.447e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1877 | 1.326e-02 | 7.167e-03 | 1.818e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3046 | 5.553e-02 | 2.560e-02 | 3.334e-01 |
|---|
| Schmidt | 0.1929 | 2.159e-02 | 2.302e-02 | 1.433e-01 |
|---|
| Sotiriou | 0.2795 | 7.763e-02 | 2.916e-02 | 4.881e-01 |
|---|
| Wang | 0.2033 | 4.514e-02 | 3.076e-02 | 1.666e-01 |
|---|
| Zhang | 0.3086 | 9.970e-03 | 5.074e-03 | 4.662e-02 |
|---|
Expression data for subnetwork 8721-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
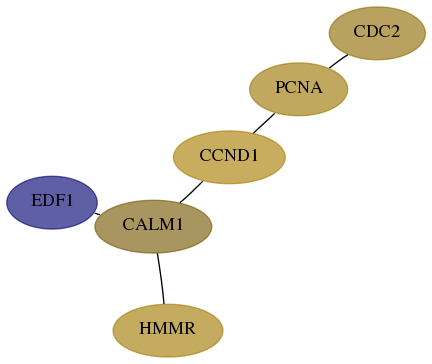
Score for each gene in subnetwork 8721-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| edf1 |   | 1 | 90 | 103 | 111 | -0.036 | 0.087 | 0.112 | 0.146 | 0.072 | 0.131 | 0.068 | 0.007 | 0.065 | 0.093 |
|---|
| hmmr |   | 10 | 14 | 35 | 23 | 0.112 | 0.310 | 0.147 | 0.127 | 0.145 | 0.221 | 0.179 | 0.233 | 0.227 | 0.144 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 8721-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 0.03783043 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 2.192E-05 | 0.02520682 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 9.354E-05 | 0.0717125 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.354E-05 | 0.05378438 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.633E-04 | 0.07509739 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.965E-04 | 0.07531495 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.142E-04 | 0.07038678 |
|---|
| base-excision repair | GO:0006284 |  | 2.142E-04 | 0.06158843 |
|---|
| response to calcium ion | GO:0051592 |  | 6.003E-04 | 0.15339937 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.307E-04 | 0.14506198 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.938E-04 | 0.14506791 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 0.02053359 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 8.405E-06 | 0.01026679 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.592E-05 | 0.02925457 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.68E-05 | 0.03469293 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.697E-05 | 0.04737849 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.127E-04 | 0.0458897 |
|---|
| base-excision repair | GO:0006284 |  | 1.127E-04 | 0.03933403 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.21E-04 | 0.03695423 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.067E-04 | 0.08325966 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.341E-04 | 0.08161178 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.341E-04 | 0.07419253 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endothelial cell differentiation | GO:0045446 |  | 9.426E-06 | 0.02268 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 0.01134 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.028E-05 | 0.03230832 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.734E-05 | 0.03449285 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.004E-04 | 0.04830826 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.174E-04 | 0.04706721 |
|---|
| base-excision repair | GO:0006284 |  | 1.174E-04 | 0.04034333 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.264E-04 | 0.03800237 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.438E-04 | 0.09190501 |
|---|
| response to calcium ion | GO:0051592 |  | 3.589E-04 | 0.08636013 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.589E-04 | 0.07850921 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 0.02053359 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 8.405E-06 | 0.01026679 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.592E-05 | 0.02925457 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.68E-05 | 0.03469293 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.697E-05 | 0.04737849 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.127E-04 | 0.0458897 |
|---|
| base-excision repair | GO:0006284 |  | 1.127E-04 | 0.03933403 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.21E-04 | 0.03695423 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.067E-04 | 0.08325966 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.341E-04 | 0.08161178 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.341E-04 | 0.07419253 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endothelial cell differentiation | GO:0045446 |  | 9.426E-06 | 0.02268 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 0.01134 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.028E-05 | 0.03230832 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.734E-05 | 0.03449285 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.004E-04 | 0.04830826 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.174E-04 | 0.04706721 |
|---|
| base-excision repair | GO:0006284 |  | 1.174E-04 | 0.04034333 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.264E-04 | 0.03800237 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.438E-04 | 0.09190501 |
|---|
| response to calcium ion | GO:0051592 |  | 3.589E-04 | 0.08636013 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.589E-04 | 0.07850921 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endothelial cell differentiation | GO:0045446 |  | 9.426E-06 | 0.02268 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 0.01134 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.028E-05 | 0.03230832 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.734E-05 | 0.03449285 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.004E-04 | 0.04830826 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.174E-04 | 0.04706721 |
|---|
| base-excision repair | GO:0006284 |  | 1.174E-04 | 0.04034333 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.264E-04 | 0.03800237 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.438E-04 | 0.09190501 |
|---|
| response to calcium ion | GO:0051592 |  | 3.589E-04 | 0.08636013 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 3.589E-04 | 0.07850921 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 0.03783043 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 2.192E-05 | 0.02520682 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 9.354E-05 | 0.0717125 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.354E-05 | 0.05378438 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.633E-04 | 0.07509739 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.965E-04 | 0.07531495 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.142E-04 | 0.07038678 |
|---|
| base-excision repair | GO:0006284 |  | 2.142E-04 | 0.06158843 |
|---|
| response to calcium ion | GO:0051592 |  | 6.003E-04 | 0.15339937 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.307E-04 | 0.14506198 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.938E-04 | 0.14506791 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 0.03783043 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 2.192E-05 | 0.02520682 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 9.354E-05 | 0.0717125 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.354E-05 | 0.05378438 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.633E-04 | 0.07509739 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.965E-04 | 0.07531495 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.142E-04 | 0.07038678 |
|---|
| base-excision repair | GO:0006284 |  | 2.142E-04 | 0.06158843 |
|---|
| response to calcium ion | GO:0051592 |  | 6.003E-04 | 0.15339937 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.307E-04 | 0.14506198 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.938E-04 | 0.14506791 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 0.03783043 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 2.192E-05 | 0.02520682 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 9.354E-05 | 0.0717125 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.354E-05 | 0.05378438 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.633E-04 | 0.07509739 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.965E-04 | 0.07531495 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.142E-04 | 0.07038678 |
|---|
| base-excision repair | GO:0006284 |  | 2.142E-04 | 0.06158843 |
|---|
| response to calcium ion | GO:0051592 |  | 6.003E-04 | 0.15339937 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.307E-04 | 0.14506198 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.938E-04 | 0.14506791 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 0.03783043 |
|---|
| endothelial cell differentiation | GO:0045446 |  | 2.192E-05 | 0.02520682 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 9.354E-05 | 0.0717125 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 9.354E-05 | 0.05378438 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.633E-04 | 0.07509739 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.965E-04 | 0.07531495 |
|---|
| fat cell differentiation | GO:0045444 |  | 2.142E-04 | 0.07038678 |
|---|
| base-excision repair | GO:0006284 |  | 2.142E-04 | 0.06158843 |
|---|
| response to calcium ion | GO:0051592 |  | 6.003E-04 | 0.15339937 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 6.307E-04 | 0.14506198 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.938E-04 | 0.14506791 |
|---|



