Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 84232-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1729 | 1.705e-02 | 6.688e-03 | 8.869e-02 |
|---|
| IPC-NIBC-129 | 0.2514 | 4.839e-02 | 3.856e-02 | 2.324e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2213 | 1.155e-02 | 7.209e-03 | 1.413e-01 |
|---|
| Loi_GPL570 | 0.2755 | 1.438e-01 | 1.748e-01 | 4.855e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1887 | 9.150e-03 | 4.362e-03 | 1.201e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3225 | 1.867e-01 | 1.211e-01 | 6.991e-01 |
|---|
| Schmidt | 0.1826 | 1.268e-02 | 1.369e-02 | 8.996e-02 |
|---|
| Sotiriou | 0.3129 | 7.642e-02 | 2.850e-02 | 4.832e-01 |
|---|
| Wang | 0.1176 | 1.922e-01 | 1.878e-01 | 4.724e-01 |
|---|
| Zhang | 0.1963 | 7.372e-03 | 3.621e-03 | 3.640e-02 |
|---|
Expression data for subnetwork 84232-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
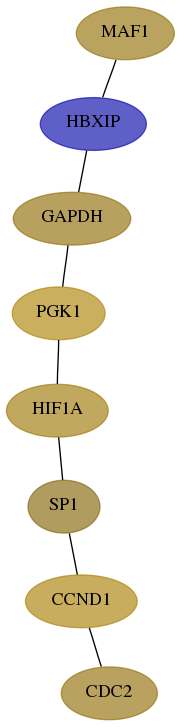
Score for each gene in subnetwork 84232-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| maf1 |   | 1 | 90 | 157 | 167 | 0.076 | -0.058 | 0.041 | 0.155 | 0.108 | 0.107 | 0.093 | 0.156 | 0.070 | 0.119 |
|---|
| pgk1 |   | 6 | 17 | 16 | 14 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.310 | 0.201 | 0.127 | 0.110 | 0.054 |
|---|
| hif1a |   | 7 | 15 | 16 | 13 | 0.097 | 0.246 | 0.089 | -0.009 | 0.101 | 0.011 | -0.043 | 0.119 | 0.172 | 0.129 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| gapdh |   | 2 | 53 | 45 | 43 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.288 | 0.064 | 0.213 | 0.056 | -0.088 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| hbxip |   | 1 | 90 | 157 | 167 | -0.128 | 0.190 | 0.074 | 0.110 | 0.123 | 0.069 | 0.104 | -0.010 | -0.076 | -0.027 |
|---|
GO Enrichment output for subnetwork 84232-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.401E-12 | 3.221E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.099E-12 | 5.864E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.473E-11 | 1.129E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.804E-11 | 1.612E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.858E-11 | 2.695E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.476E-10 | 5.66E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.283E-09 | 2.064E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.266E-08 | 1.514E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.275E-07 | 5.815E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.244E-07 | 9.761E-05 |
|---|
| viral genome replication | GO:0019079 |  | 9.584E-07 | 2.004E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 8.965E-14 | 2.19E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.098E-13 | 3.784E-10 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.04E-12 | 8.472E-10 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.247E-12 | 1.372E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.771E-12 | 2.331E-09 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.294E-09 | 5.269E-07 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.131E-08 | 7.436E-06 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 4.696E-08 | 1.434E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 8.763E-08 | 2.379E-05 |
|---|
| viral genome replication | GO:0019079 |  | 2.278E-07 | 5.564E-05 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 9.143E-07 | 2.03E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 2.103E-13 | 5.059E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.456E-13 | 8.97E-10 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.555E-12 | 2.049E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.579E-12 | 3.356E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.195E-11 | 5.753E-09 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.127E-09 | 8.529E-07 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.501E-08 | 1.203E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.714E-08 | 2.32E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.439E-07 | 3.848E-05 |
|---|
| viral genome replication | GO:0019079 |  | 3.74E-07 | 8.998E-05 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.375E-06 | 3.009E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 8.965E-14 | 2.19E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.098E-13 | 3.784E-10 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.04E-12 | 8.472E-10 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.247E-12 | 1.372E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.771E-12 | 2.331E-09 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.294E-09 | 5.269E-07 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.131E-08 | 7.436E-06 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 4.696E-08 | 1.434E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 8.763E-08 | 2.379E-05 |
|---|
| viral genome replication | GO:0019079 |  | 2.278E-07 | 5.564E-05 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 9.143E-07 | 2.03E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 2.103E-13 | 5.059E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.456E-13 | 8.97E-10 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.555E-12 | 2.049E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.579E-12 | 3.356E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.195E-11 | 5.753E-09 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.127E-09 | 8.529E-07 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.501E-08 | 1.203E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.714E-08 | 2.32E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.439E-07 | 3.848E-05 |
|---|
| viral genome replication | GO:0019079 |  | 3.74E-07 | 8.998E-05 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.375E-06 | 3.009E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 2.103E-13 | 5.059E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.456E-13 | 8.97E-10 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.555E-12 | 2.049E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.579E-12 | 3.356E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.195E-11 | 5.753E-09 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.127E-09 | 8.529E-07 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.501E-08 | 1.203E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.714E-08 | 2.32E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.439E-07 | 3.848E-05 |
|---|
| viral genome replication | GO:0019079 |  | 3.74E-07 | 8.998E-05 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.375E-06 | 3.009E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.401E-12 | 3.221E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.099E-12 | 5.864E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.473E-11 | 1.129E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.804E-11 | 1.612E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.858E-11 | 2.695E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.476E-10 | 5.66E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.283E-09 | 2.064E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.266E-08 | 1.514E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.275E-07 | 5.815E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.244E-07 | 9.761E-05 |
|---|
| viral genome replication | GO:0019079 |  | 9.584E-07 | 2.004E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.401E-12 | 3.221E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.099E-12 | 5.864E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.473E-11 | 1.129E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.804E-11 | 1.612E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.858E-11 | 2.695E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.476E-10 | 5.66E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.283E-09 | 2.064E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.266E-08 | 1.514E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.275E-07 | 5.815E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.244E-07 | 9.761E-05 |
|---|
| viral genome replication | GO:0019079 |  | 9.584E-07 | 2.004E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.401E-12 | 3.221E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.099E-12 | 5.864E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.473E-11 | 1.129E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.804E-11 | 1.612E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.858E-11 | 2.695E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.476E-10 | 5.66E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.283E-09 | 2.064E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.266E-08 | 1.514E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.275E-07 | 5.815E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.244E-07 | 9.761E-05 |
|---|
| viral genome replication | GO:0019079 |  | 9.584E-07 | 2.004E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.401E-12 | 3.221E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.099E-12 | 5.864E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.473E-11 | 1.129E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.804E-11 | 1.612E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.858E-11 | 2.695E-08 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.476E-10 | 5.66E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.283E-09 | 2.064E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 5.266E-08 | 1.514E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.275E-07 | 5.815E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.244E-07 | 9.761E-05 |
|---|
| viral genome replication | GO:0019079 |  | 9.584E-07 | 2.004E-04 |
|---|



