Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 79980-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.0667 | 1.458e-02 | 5.405e-03 | 7.846e-02 |
|---|
| IPC-NIBC-129 | 0.2457 | 5.268e-02 | 4.248e-02 | 2.513e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2288 | 1.295e-02 | 8.246e-03 | 1.560e-01 |
|---|
| Loi_GPL570 | 0.2689 | 7.035e-02 | 9.075e-02 | 2.931e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1815 | 3.692e-01 | 4.532e-01 | 9.967e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3027 | 1.313e-01 | 7.719e-02 | 5.825e-01 |
|---|
| Schmidt | 0.2371 | 2.780e-02 | 2.947e-02 | 1.772e-01 |
|---|
| Sotiriou | 0.2636 | 8.570e-02 | 3.358e-02 | 5.187e-01 |
|---|
| Wang | 0.1669 | 8.419e-02 | 6.785e-02 | 2.662e-01 |
|---|
| Zhang | 0.2295 | 1.029e-02 | 5.257e-03 | 4.784e-02 |
|---|
Expression data for subnetwork 79980-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
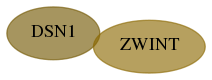
Score for each gene in subnetwork 79980-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| zwint |   | 1 | 90 | 204 | 204 | 0.067 | 0.265 | 0.232 | 0.265 | 0.191 | 0.275 | 0.232 | 0.219 | 0.162 | 0.247 |
|---|
| dsn1 |   | 1 | 90 | 204 | 204 | 0.037 | 0.097 | 0.100 | 0.214 | 0.139 | 0.247 | 0.182 | 0.247 | 0.120 | 0.089 |
|---|
GO Enrichment output for subnetwork 79980-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 2.847E-06 | 3.274E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.246E-06 | 2.489E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 3.455E-06 | 1.987E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 3.671E-06 | 1.689E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.319E-05 | 5.058E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.626E-05 | 5.344E-03 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.281E-05 | 6.559E-03 |
|---|
| regulation of interferon-alpha biosynthetic process | GO:0045354 |  | 5.72E-04 | 0.14616538 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.72E-04 | 0.13154884 |
|---|
| positive regulation of G-protein coupled receptor protein signaling pathway | GO:0045745 |  | 5.72E-04 | 0.11958986 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 7.852E-07 | 9.591E-04 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 9.974E-07 | 8.122E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.054E-06 | 6.439E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.173E-06 | 5.732E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.934E-06 | 1.602E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 4.393E-06 | 1.533E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.813E-04 | 0.08590981 |
|---|
| negative regulation of nucleotide biosynthetic process | GO:0030809 |  | 2.813E-04 | 0.07636427 |
|---|
| positive regulation of muscle development | GO:0048636 |  | 2.813E-04 | 0.06872785 |
|---|
| positive regulation of interferon-alpha biosynthetic process | GO:0045356 |  | 2.813E-04 | 0.06247986 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 1.094E-06 | 1.316E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.389E-06 | 1.114E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.469E-06 | 8.833E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.55E-06 | 7.459E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.173E-06 | 2.074E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 6.119E-06 | 2.103E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.32E-04 | 0.09985391 |
|---|
| negative regulation of nucleotide biosynthetic process | GO:0030809 |  | 3.32E-04 | 0.08875903 |
|---|
| positive regulation of interferon-alpha biosynthetic process | GO:0045356 |  | 3.32E-04 | 0.07988313 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 3.32E-04 | 0.07262103 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 7.852E-07 | 9.591E-04 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 9.974E-07 | 8.122E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.054E-06 | 6.439E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.173E-06 | 5.732E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.934E-06 | 1.602E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 4.393E-06 | 1.533E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.813E-04 | 0.08590981 |
|---|
| negative regulation of nucleotide biosynthetic process | GO:0030809 |  | 2.813E-04 | 0.07636427 |
|---|
| positive regulation of muscle development | GO:0048636 |  | 2.813E-04 | 0.06872785 |
|---|
| positive regulation of interferon-alpha biosynthetic process | GO:0045356 |  | 2.813E-04 | 0.06247986 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 1.094E-06 | 1.316E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.389E-06 | 1.114E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.469E-06 | 8.833E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.55E-06 | 7.459E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.173E-06 | 2.074E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 6.119E-06 | 2.103E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.32E-04 | 0.09985391 |
|---|
| negative regulation of nucleotide biosynthetic process | GO:0030809 |  | 3.32E-04 | 0.08875903 |
|---|
| positive regulation of interferon-alpha biosynthetic process | GO:0045356 |  | 3.32E-04 | 0.07988313 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 3.32E-04 | 0.07262103 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 1.094E-06 | 1.316E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.389E-06 | 1.114E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.469E-06 | 8.833E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.55E-06 | 7.459E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.173E-06 | 2.074E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 6.119E-06 | 2.103E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.32E-04 | 0.09985391 |
|---|
| negative regulation of nucleotide biosynthetic process | GO:0030809 |  | 3.32E-04 | 0.08875903 |
|---|
| positive regulation of interferon-alpha biosynthetic process | GO:0045356 |  | 3.32E-04 | 0.07988313 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 3.32E-04 | 0.07262103 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 2.847E-06 | 3.274E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.246E-06 | 2.489E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 3.455E-06 | 1.987E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 3.671E-06 | 1.689E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.319E-05 | 5.058E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.626E-05 | 5.344E-03 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.281E-05 | 6.559E-03 |
|---|
| regulation of interferon-alpha biosynthetic process | GO:0045354 |  | 5.72E-04 | 0.14616538 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.72E-04 | 0.13154884 |
|---|
| positive regulation of G-protein coupled receptor protein signaling pathway | GO:0045745 |  | 5.72E-04 | 0.11958986 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 2.847E-06 | 3.274E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.246E-06 | 2.489E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 3.455E-06 | 1.987E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 3.671E-06 | 1.689E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.319E-05 | 5.058E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.626E-05 | 5.344E-03 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.281E-05 | 6.559E-03 |
|---|
| regulation of interferon-alpha biosynthetic process | GO:0045354 |  | 5.72E-04 | 0.14616538 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.72E-04 | 0.13154884 |
|---|
| positive regulation of G-protein coupled receptor protein signaling pathway | GO:0045745 |  | 5.72E-04 | 0.11958986 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 2.847E-06 | 3.274E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.246E-06 | 2.489E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 3.455E-06 | 1.987E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 3.671E-06 | 1.689E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.319E-05 | 5.058E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.626E-05 | 5.344E-03 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.281E-05 | 6.559E-03 |
|---|
| regulation of interferon-alpha biosynthetic process | GO:0045354 |  | 5.72E-04 | 0.14616538 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.72E-04 | 0.13154884 |
|---|
| positive regulation of G-protein coupled receptor protein signaling pathway | GO:0045745 |  | 5.72E-04 | 0.11958986 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 0E00 | 0E00 |
|---|
| spindle organization | GO:0007051 |  | 2.847E-06 | 3.274E-03 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.246E-06 | 2.489E-03 |
|---|
| sister chromatid segregation | GO:0000819 |  | 3.455E-06 | 1.987E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 3.671E-06 | 1.689E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.319E-05 | 5.058E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.626E-05 | 5.344E-03 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.281E-05 | 6.559E-03 |
|---|
| regulation of interferon-alpha biosynthetic process | GO:0045354 |  | 5.72E-04 | 0.14616538 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.72E-04 | 0.13154884 |
|---|
| positive regulation of G-protein coupled receptor protein signaling pathway | GO:0045745 |  | 5.72E-04 | 0.11958986 |
|---|



