Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7916-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1672 | 1.140e-02 | 3.866e-03 | 6.463e-02 |
|---|
| IPC-NIBC-129 | 0.2117 | 3.591e-02 | 2.743e-02 | 1.747e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2405 | 2.548e-02 | 1.824e-02 | 2.710e-01 |
|---|
| Loi_GPL570 | 0.2989 | 6.814e-02 | 8.814e-02 | 2.861e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1801 | 1.145e-02 | 5.890e-03 | 1.549e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3959 | 7.665e-02 | 3.871e-02 | 4.176e-01 |
|---|
| Schmidt | 0.2394 | 2.011e-02 | 2.148e-02 | 1.348e-01 |
|---|
| Sotiriou | 0.2840 | 8.770e-02 | 3.470e-02 | 5.260e-01 |
|---|
| Wang | 0.1959 | 5.125e-02 | 3.618e-02 | 1.836e-01 |
|---|
| Zhang | 0.2793 | 2.113e-03 | 8.990e-04 | 1.282e-02 |
|---|
Expression data for subnetwork 7916-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
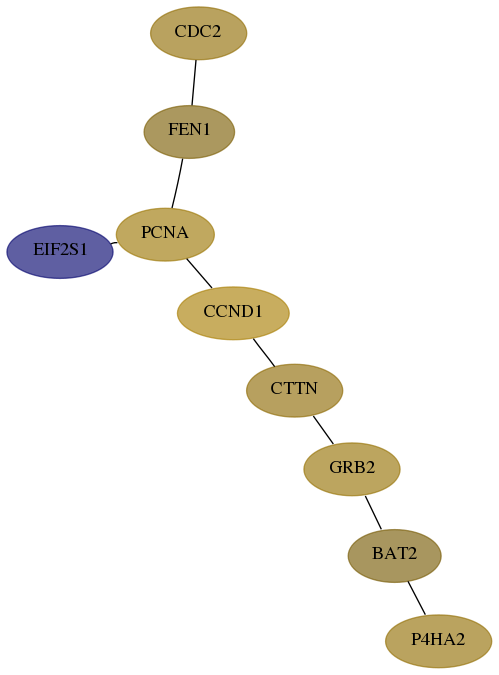
Score for each gene in subnetwork 7916-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| fen1 |   | 7 | 15 | 8 | 6 | 0.045 | 0.194 | 0.169 | 0.196 | 0.136 | 0.332 | 0.221 | 0.144 | 0.106 | 0.056 |
|---|
| cttn |   | 5 | 20 | 8 | 9 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.216 | 0.069 | 0.115 | 0.129 | 0.025 |
|---|
| bat2 |   | 1 | 90 | 8 | 33 | 0.039 | 0.182 | -0.001 | -0.013 | 0.062 | 0.109 | 0.186 | 0.099 | -0.078 | 0.059 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| p4ha2 |   | 1 | 90 | 8 | 33 | 0.077 | 0.313 | 0.070 | 0.159 | 0.149 | 0.180 | 0.055 | 0.105 | 0.184 | 0.043 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
| grb2 |   | 6 | 17 | 8 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 |
|---|
GO Enrichment output for subnetwork 7916-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 0.03689356 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.02693876 |
|---|
| UV protection | GO:0009650 |  | 5.01E-05 | 0.03840915 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.03413632 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.06121606 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.331E-04 | 0.05101339 |
|---|
| response to light stimulus | GO:0009416 |  | 1.459E-04 | 0.04793355 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.321E-04 | 0.06673843 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.793E-04 | 0.07137757 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.045E-04 | 0.07003439 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.045E-04 | 0.06366763 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 3.923E-06 | 9.583E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 0.01026679 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.002E-05 | 8.163E-03 |
|---|
| UV protection | GO:0009650 |  | 1.649E-05 | 0.01007325 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.592E-05 | 0.01755274 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.68E-05 | 0.02312862 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.697E-05 | 0.03384178 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.047E-04 | 0.03196969 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.127E-04 | 0.03059313 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.127E-04 | 0.02753382 |
|---|
| base-excision repair | GO:0006284 |  | 1.127E-04 | 0.02503074 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 5.743E-06 | 0.01381855 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 0.01407861 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.64E-05 | 0.01315413 |
|---|
| UV protection | GO:0009650 |  | 2.296E-05 | 0.01381069 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5E-05 | 0.02405795 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.116E-05 | 0.02853469 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.245E-04 | 0.04280687 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.349E-04 | 0.04056119 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.456E-04 | 0.03892323 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.456E-04 | 0.03503091 |
|---|
| base-excision repair | GO:0006284 |  | 1.456E-04 | 0.03184628 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 3.923E-06 | 9.583E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 0.01026679 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.002E-05 | 8.163E-03 |
|---|
| UV protection | GO:0009650 |  | 1.649E-05 | 0.01007325 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.592E-05 | 0.01755274 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.68E-05 | 0.02312862 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.697E-05 | 0.03384178 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.047E-04 | 0.03196969 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.127E-04 | 0.03059313 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.127E-04 | 0.02753382 |
|---|
| base-excision repair | GO:0006284 |  | 1.127E-04 | 0.02503074 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 5.743E-06 | 0.01381855 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 0.01407861 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.64E-05 | 0.01315413 |
|---|
| UV protection | GO:0009650 |  | 2.296E-05 | 0.01381069 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5E-05 | 0.02405795 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.116E-05 | 0.02853469 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.245E-04 | 0.04280687 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.349E-04 | 0.04056119 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.456E-04 | 0.03892323 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.456E-04 | 0.03503091 |
|---|
| base-excision repair | GO:0006284 |  | 1.456E-04 | 0.03184628 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 5.743E-06 | 0.01381855 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 0.01407861 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.64E-05 | 0.01315413 |
|---|
| UV protection | GO:0009650 |  | 2.296E-05 | 0.01381069 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5E-05 | 0.02405795 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.116E-05 | 0.02853469 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.245E-04 | 0.04280687 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.349E-04 | 0.04056119 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.456E-04 | 0.03892323 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.456E-04 | 0.03503091 |
|---|
| base-excision repair | GO:0006284 |  | 1.456E-04 | 0.03184628 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 0.03689356 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.02693876 |
|---|
| UV protection | GO:0009650 |  | 5.01E-05 | 0.03840915 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.03413632 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.06121606 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.331E-04 | 0.05101339 |
|---|
| response to light stimulus | GO:0009416 |  | 1.459E-04 | 0.04793355 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.321E-04 | 0.06673843 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.793E-04 | 0.07137757 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.045E-04 | 0.07003439 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.045E-04 | 0.06366763 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 0.03689356 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.02693876 |
|---|
| UV protection | GO:0009650 |  | 5.01E-05 | 0.03840915 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.03413632 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.06121606 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.331E-04 | 0.05101339 |
|---|
| response to light stimulus | GO:0009416 |  | 1.459E-04 | 0.04793355 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.321E-04 | 0.06673843 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.793E-04 | 0.07137757 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.045E-04 | 0.07003439 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.045E-04 | 0.06366763 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 0.03689356 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.02693876 |
|---|
| UV protection | GO:0009650 |  | 5.01E-05 | 0.03840915 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.03413632 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.06121606 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.331E-04 | 0.05101339 |
|---|
| response to light stimulus | GO:0009416 |  | 1.459E-04 | 0.04793355 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.321E-04 | 0.06673843 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.793E-04 | 0.07137757 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.045E-04 | 0.07003439 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.045E-04 | 0.06366763 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 0.03689356 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.02693876 |
|---|
| UV protection | GO:0009650 |  | 5.01E-05 | 0.03840915 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.03413632 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.06121606 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.331E-04 | 0.05101339 |
|---|
| response to light stimulus | GO:0009416 |  | 1.459E-04 | 0.04793355 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.321E-04 | 0.06673843 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.793E-04 | 0.07137757 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.045E-04 | 0.07003439 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.045E-04 | 0.06366763 |
|---|



