Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7528-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1458 | 1.196e-02 | 4.127e-03 | 6.713e-02 |
|---|
| IPC-NIBC-129 | 0.1825 | 5.787e-02 | 4.728e-02 | 2.736e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2383 | 4.523e-02 | 3.562e-02 | 4.088e-01 |
|---|
| Loi_GPL570 | 0.2614 | 9.751e-02 | 1.225e-01 | 3.725e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1911 | 2.606e-02 | 1.754e-02 | 3.501e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3024 | 9.057e-02 | 4.794e-02 | 4.659e-01 |
|---|
| Schmidt | 0.2124 | 3.238e-02 | 3.421e-02 | 2.007e-01 |
|---|
| Sotiriou | 0.2539 | 7.345e-02 | 2.694e-02 | 4.712e-01 |
|---|
| Wang | 0.2275 | 2.969e-02 | 1.795e-02 | 1.201e-01 |
|---|
| Zhang | 0.2640 | 1.034e-02 | 5.286e-03 | 4.804e-02 |
|---|
Expression data for subnetwork 7528-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
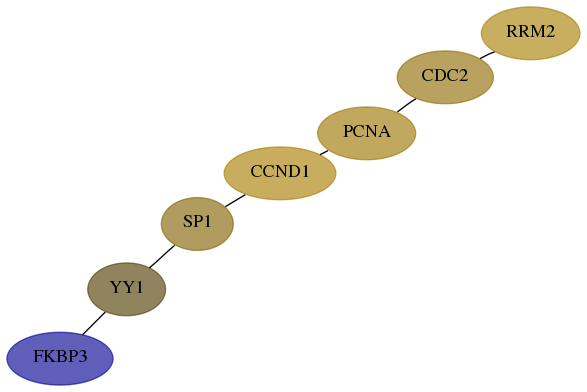
Score for each gene in subnetwork 7528-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| rrm2 |   | 5 | 20 | 35 | 25 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 |
|---|
| fkbp3 |   | 2 | 53 | 129 | 116 | -0.072 | 0.129 | 0.144 | 0.166 | 0.136 | 0.010 | 0.159 | 0.052 | 0.058 | 0.199 |
|---|
| yy1 |   | 1 | 90 | 129 | 135 | 0.016 | 0.002 | 0.109 | 0.146 | 0.070 | -0.005 | 0.040 | 0.059 | 0.159 | 0.085 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 7528-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.416E-09 | 1.476E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.122E-08 | 1.291E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.239E-07 | 3.25E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 2.574E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.366E-05 | 5.237E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.331E-05 | 0.01751529 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.09E-05 | 0.01750794 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.09E-05 | 0.01556262 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 7.758E-05 | 0.01784312 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 9.626E-05 | 0.02012686 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.04E-06 | 2.54E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.661E-06 | 2.029E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.491E-06 | 2.028E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.486E-06 | 2.129E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.648E-06 | 2.271E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.974E-06 | 2.432E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.988E-05 | 6.938E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.252E-05 | 6.878E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.144E-05 | 8.535E-03 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 3.475E-05 | 8.488E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 3.821E-05 | 8.486E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.3E-09 | 1.035E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.031E-08 | 1.241E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.883E-07 | 1.51E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.097E-06 | 1.261E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.644E-06 | 1.272E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.964E-06 | 1.59E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 2.542E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 9.134E-06 | 2.747E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.162E-05 | 8.452E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.582E-05 | 8.618E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 9.845E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.04E-06 | 2.54E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.661E-06 | 2.029E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.491E-06 | 2.028E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.486E-06 | 2.129E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.648E-06 | 2.271E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.974E-06 | 2.432E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.988E-05 | 6.938E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.252E-05 | 6.878E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.144E-05 | 8.535E-03 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 3.475E-05 | 8.488E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 3.821E-05 | 8.486E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.3E-09 | 1.035E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.031E-08 | 1.241E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.883E-07 | 1.51E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.097E-06 | 1.261E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.644E-06 | 1.272E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.964E-06 | 1.59E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 2.542E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 9.134E-06 | 2.747E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.162E-05 | 8.452E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.582E-05 | 8.618E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 9.845E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.3E-09 | 1.035E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.031E-08 | 1.241E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.883E-07 | 1.51E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.097E-06 | 1.261E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.644E-06 | 1.272E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.964E-06 | 1.59E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 2.542E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 9.134E-06 | 2.747E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.162E-05 | 8.452E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.582E-05 | 8.618E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 9.845E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.416E-09 | 1.476E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.122E-08 | 1.291E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.239E-07 | 3.25E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 2.574E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.366E-05 | 5.237E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.331E-05 | 0.01751529 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.09E-05 | 0.01750794 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.09E-05 | 0.01556262 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 7.758E-05 | 0.01784312 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 9.626E-05 | 0.02012686 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.416E-09 | 1.476E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.122E-08 | 1.291E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.239E-07 | 3.25E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 2.574E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.366E-05 | 5.237E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.331E-05 | 0.01751529 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.09E-05 | 0.01750794 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.09E-05 | 0.01556262 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 7.758E-05 | 0.01784312 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 9.626E-05 | 0.02012686 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.416E-09 | 1.476E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.122E-08 | 1.291E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.239E-07 | 3.25E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 2.574E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.366E-05 | 5.237E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.331E-05 | 0.01751529 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.09E-05 | 0.01750794 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.09E-05 | 0.01556262 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 7.758E-05 | 0.01784312 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 9.626E-05 | 0.02012686 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.416E-09 | 1.476E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.122E-08 | 1.291E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.239E-07 | 3.25E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 2.574E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.366E-05 | 5.237E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.331E-05 | 0.01751529 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.09E-05 | 0.01750794 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.09E-05 | 0.01556262 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 7.758E-05 | 0.01784312 |
|---|
| camera-type eye morphogenesis | GO:0048593 |  | 9.626E-05 | 0.02012686 |
|---|



