Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7374-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1489 | 2.567e-02 | 1.165e-02 | 1.216e-01 |
|---|
| IPC-NIBC-129 | 0.2004 | 8.563e-02 | 7.377e-02 | 3.823e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2016 | 3.179e-02 | 2.363e-02 | 3.198e-01 |
|---|
| Loi_GPL570 | 0.2303 | 1.163e-01 | 1.440e-01 | 4.215e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1820 | 2.320e-02 | 1.505e-02 | 3.159e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3364 | 1.486e-01 | 9.044e-02 | 6.232e-01 |
|---|
| Schmidt | 0.1989 | 2.863e-02 | 3.033e-02 | 1.815e-01 |
|---|
| Sotiriou | 0.2617 | 8.510e-02 | 3.324e-02 | 5.165e-01 |
|---|
| Wang | 0.2001 | 4.770e-02 | 3.301e-02 | 1.738e-01 |
|---|
| Zhang | 0.2179 | 5.824e-03 | 2.784e-03 | 2.996e-02 |
|---|
Expression data for subnetwork 7374-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
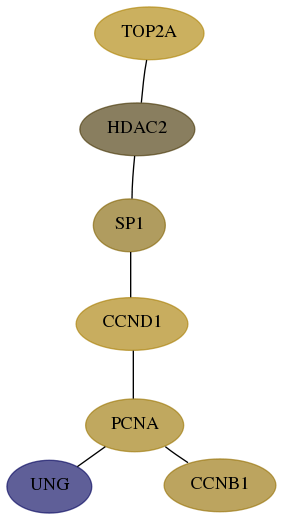
Score for each gene in subnetwork 7374-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| hdac2 |   | 16 | 10 | 64 | 39 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 |
|---|
| ung |   | 1 | 90 | 209 | 211 | -0.021 | 0.153 | 0.118 | 0.098 | 0.118 | 0.160 | 0.003 | 0.246 | 0.094 | 0.021 |
|---|
| top2a |   | 22 | 7 | 70 | 46 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 |
|---|
| ccnb1 |   | 33 | 4 | 68 | 40 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 7374-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 6.235E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.942E-06 | 2.256E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.942E-06 | 1.692E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 3.88E-06 | 1.785E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.412E-06 | 1.691E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.029E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 2.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.058E-05 | 2.704E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.322E-05 | 3.041E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 2.813E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 9.585E-08 | 2.342E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.041E-07 | 6.158E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.044E-06 | 8.504E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.044E-06 | 6.378E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 1.391E-06 | 6.798E-04 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.566E-06 | 6.377E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.566E-06 | 5.466E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.958E-06 | 5.98E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 7.932E-04 |
|---|
| DNA topological change | GO:0006265 |  | 2.922E-06 | 7.139E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.391E-06 | 9.753E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 1.406E-07 | 3.383E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 9.954E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.454E-06 | 1.166E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.454E-06 | 8.748E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.454E-06 | 6.998E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.181E-06 | 8.746E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 2.282E-06 | 7.844E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.211E-06 | 9.657E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.088E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.07E-06 | 9.791E-04 |
|---|
| DNA ligation | GO:0006266 |  | 6.537E-06 | 1.43E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 9.585E-08 | 2.342E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.041E-07 | 6.158E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.044E-06 | 8.504E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.044E-06 | 6.378E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 1.391E-06 | 6.798E-04 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.566E-06 | 6.377E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.566E-06 | 5.466E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.958E-06 | 5.98E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 7.932E-04 |
|---|
| DNA topological change | GO:0006265 |  | 2.922E-06 | 7.139E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.391E-06 | 9.753E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 1.406E-07 | 3.383E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 9.954E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.454E-06 | 1.166E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.454E-06 | 8.748E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.454E-06 | 6.998E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.181E-06 | 8.746E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 2.282E-06 | 7.844E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.211E-06 | 9.657E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.088E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.07E-06 | 9.791E-04 |
|---|
| DNA ligation | GO:0006266 |  | 6.537E-06 | 1.43E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 1.406E-07 | 3.383E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.275E-07 | 9.954E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.454E-06 | 1.166E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.454E-06 | 8.748E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.454E-06 | 6.998E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.181E-06 | 8.746E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 2.282E-06 | 7.844E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.211E-06 | 9.657E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.07E-06 | 1.088E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.07E-06 | 9.791E-04 |
|---|
| DNA ligation | GO:0006266 |  | 6.537E-06 | 1.43E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 6.235E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.942E-06 | 2.256E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.942E-06 | 1.692E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 3.88E-06 | 1.785E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.412E-06 | 1.691E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.029E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 2.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.058E-05 | 2.704E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.322E-05 | 3.041E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 2.813E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 6.235E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.942E-06 | 2.256E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.942E-06 | 1.692E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 3.88E-06 | 1.785E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.412E-06 | 1.691E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.029E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 2.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.058E-05 | 2.704E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.322E-05 | 3.041E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 2.813E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 6.235E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.942E-06 | 2.256E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.942E-06 | 1.692E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 3.88E-06 | 1.785E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.412E-06 | 1.691E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.029E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 2.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.058E-05 | 2.704E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.322E-05 | 3.041E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 2.813E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair | GO:0006284 |  | 2.711E-07 | 6.235E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.942E-06 | 2.256E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.942E-06 | 1.692E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 3.88E-06 | 1.785E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.412E-06 | 1.691E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.029E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 2.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.058E-05 | 2.704E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.322E-05 | 3.041E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.345E-05 | 2.813E-03 |
|---|



