Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5983-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1698 | 9.906e-03 | 3.191e-03 | 5.780e-02 |
|---|
| IPC-NIBC-129 | 0.1924 | 1.185e-01 | 1.064e-01 | 4.904e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2472 | 3.724e-02 | 2.841e-02 | 3.580e-01 |
|---|
| Loi_GPL570 | 0.2042 | 5.110e-02 | 6.765e-02 | 2.288e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1570 | 1.037e-02 | 5.157e-03 | 1.386e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3315 | 9.726e-02 | 5.253e-02 | 4.873e-01 |
|---|
| Schmidt | 0.2609 | 4.472e-02 | 4.691e-02 | 2.591e-01 |
|---|
| Sotiriou | 0.2333 | 1.268e-01 | 5.867e-02 | 6.455e-01 |
|---|
| Wang | 0.2096 | 4.046e-02 | 2.673e-02 | 1.531e-01 |
|---|
| Zhang | 0.2574 | 6.327e-03 | 3.053e-03 | 3.209e-02 |
|---|
Expression data for subnetwork 5983-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
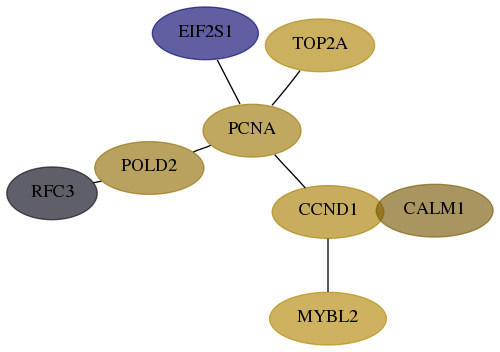
Score for each gene in subnetwork 5983-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pold2 |   | 3 | 39 | 89 | 70 | 0.073 | 0.180 | 0.204 | 0.131 | 0.150 | 0.186 | 0.146 | 0.126 | -0.084 | 0.072 |
|---|
| rfc3 |   | 1 | 90 | 133 | 138 | -0.002 | 0.231 | 0.169 | -0.019 | 0.074 | 0.190 | 0.193 | 0.114 | 0.133 | 0.027 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| mybl2 |   | 5 | 20 | 92 | 63 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.275 | 0.202 | 0.182 | 0.120 | 0.074 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| top2a |   | 22 | 7 | 70 | 46 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
GO Enrichment output for subnetwork 5983-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.798E-09 | 4.136E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.61E-07 | 3.001E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 6.422E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 7.702E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.121E-05 | 0.01025573 |
|---|
| DNA ligation | GO:0006266 |  | 4.011E-05 | 0.01153022 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.01E-05 | 0.01280305 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.01365453 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.02782548 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.893E-10 | 7.067E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 6.164E-08 | 7.529E-05 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.364E-06 | 3.554E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.543E-06 | 3.996E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 5.963E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.22E-05 | 4.969E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.568E-05 | 5.474E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.769E-05 | 5.402E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.96E-05 | 5.32E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.96E-05 | 4.788E-03 |
|---|
| regulation of viral genome replication | GO:0045069 |  | 6.641E-05 | 0.01474937 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.607E-10 | 1.349E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.108E-07 | 1.333E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.077E-06 | 4.874E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.077E-06 | 3.655E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 8.176E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.699E-05 | 6.813E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 2.183E-05 | 7.505E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.728E-05 | 8.205E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.728E-05 | 7.293E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 2.891E-05 | 6.956E-03 |
|---|
| regulation of viral genome replication | GO:0045069 |  | 8.217E-05 | 0.01797212 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.893E-10 | 7.067E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 6.164E-08 | 7.529E-05 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.364E-06 | 3.554E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.543E-06 | 3.996E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 5.963E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.22E-05 | 4.969E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.568E-05 | 5.474E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.769E-05 | 5.402E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.96E-05 | 5.32E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.96E-05 | 4.788E-03 |
|---|
| regulation of viral genome replication | GO:0045069 |  | 6.641E-05 | 0.01474937 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.607E-10 | 1.349E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.108E-07 | 1.333E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.077E-06 | 4.874E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.077E-06 | 3.655E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 8.176E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.699E-05 | 6.813E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 2.183E-05 | 7.505E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.728E-05 | 8.205E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.728E-05 | 7.293E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 2.891E-05 | 6.956E-03 |
|---|
| regulation of viral genome replication | GO:0045069 |  | 8.217E-05 | 0.01797212 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.607E-10 | 1.349E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.108E-07 | 1.333E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.077E-06 | 4.874E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.077E-06 | 3.655E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 8.176E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.699E-05 | 6.813E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 2.183E-05 | 7.505E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.728E-05 | 8.205E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.728E-05 | 7.293E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 2.891E-05 | 6.956E-03 |
|---|
| regulation of viral genome replication | GO:0045069 |  | 8.217E-05 | 0.01797212 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.798E-09 | 4.136E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.61E-07 | 3.001E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 6.422E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 7.702E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.121E-05 | 0.01025573 |
|---|
| DNA ligation | GO:0006266 |  | 4.011E-05 | 0.01153022 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.01E-05 | 0.01280305 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.01365453 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.02782548 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.798E-09 | 4.136E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.61E-07 | 3.001E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 6.422E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 7.702E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.121E-05 | 0.01025573 |
|---|
| DNA ligation | GO:0006266 |  | 4.011E-05 | 0.01153022 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.01E-05 | 0.01280305 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.01365453 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.02782548 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.798E-09 | 4.136E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.61E-07 | 3.001E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 6.422E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 7.702E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.121E-05 | 0.01025573 |
|---|
| DNA ligation | GO:0006266 |  | 4.011E-05 | 0.01153022 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.01E-05 | 0.01280305 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.01365453 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.02782548 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.798E-09 | 4.136E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.61E-07 | 3.001E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 6.422E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 7.702E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 8.98E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.121E-05 | 0.01025573 |
|---|
| DNA ligation | GO:0006266 |  | 4.011E-05 | 0.01153022 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.01E-05 | 0.01280305 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 5.937E-05 | 0.01365453 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.331E-04 | 0.02782548 |
|---|



