Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5460-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1094 | 1.564e-02 | 5.950e-03 | 8.293e-02 |
|---|
| IPC-NIBC-129 | 0.2006 | 3.409e-02 | 2.584e-02 | 1.659e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2254 | 3.167e-02 | 2.352e-02 | 3.188e-01 |
|---|
| Loi_GPL570 | 0.3029 | 1.361e-01 | 1.662e-01 | 4.683e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1878 | 9.706e-02 | 9.476e-02 | 8.009e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3283 | 1.166e-01 | 6.625e-02 | 5.439e-01 |
|---|
| Schmidt | 0.1869 | 3.036e-02 | 3.213e-02 | 1.905e-01 |
|---|
| Sotiriou | 0.2580 | 7.747e-02 | 2.907e-02 | 4.874e-01 |
|---|
| Wang | 0.2053 | 4.362e-02 | 2.944e-02 | 1.622e-01 |
|---|
| Zhang | 0.2406 | 6.677e-03 | 3.242e-03 | 3.355e-02 |
|---|
Expression data for subnetwork 5460-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
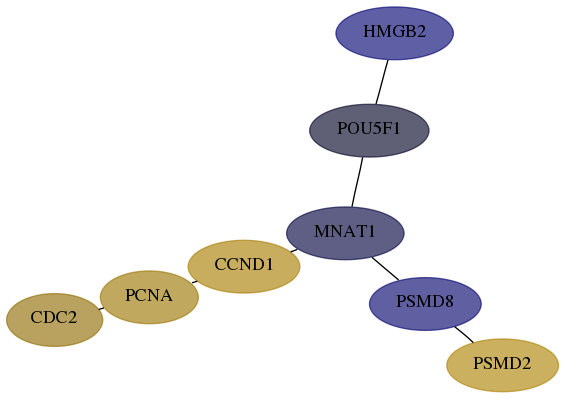
Score for each gene in subnetwork 5460-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| psmd2 |   | 3 | 39 | 20 | 22 | 0.147 | 0.322 | 0.122 | 0.146 | 0.143 | 0.194 | 0.318 | 0.151 | 0.154 | 0.042 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| pou5f1 |   | 1 | 90 | 153 | 160 | -0.005 | 0.152 | 0.070 | 0.087 | 0.074 | 0.082 | 0.018 | 0.016 | 0.066 | 0.058 |
|---|
| mnat1 |   | 1 | 90 | 153 | 160 | -0.011 | 0.083 | 0.075 | 0.305 | 0.105 | 0.107 | 0.118 | -0.002 | 0.144 | 0.307 |
|---|
| hmgb2 |   | 1 | 90 | 153 | 160 | -0.035 | 0.199 | 0.149 | 0.166 | 0.180 | 0.172 | 0.210 | 0.177 | 0.096 | 0.138 |
|---|
| psmd8 |   | 1 | 90 | 153 | 160 | -0.031 | 0.158 | 0.163 | 0.198 | 0.168 | 0.274 | -0.009 | 0.177 | 0.086 | -0.069 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 5460-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.346E-08 | 9.996E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.645E-08 | 5.341E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.632E-08 | 4.318E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.632E-08 | 3.238E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.182E-08 | 3.304E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.066E-08 | 3.092E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.008E-07 | 3.311E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.309E-07 | 3.764E-05 |
|---|
| base-excision repair | GO:0006284 |  | 3.724E-07 | 9.517E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.808E-06 | 4.158E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 7.938E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.208E-08 | 2.952E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.283E-08 | 1.567E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.525E-08 | 1.242E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.525E-08 | 9.315E-06 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.004E-08 | 9.792E-06 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.224E-08 | 9.055E-06 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.853E-08 | 9.956E-06 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 4.308E-08 | 1.316E-05 |
|---|
| base-excision repair | GO:0006284 |  | 1.584E-07 | 4.3E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 8.866E-07 | 2.166E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.405E-06 | 3.121E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.673E-08 | 4.026E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.776E-08 | 2.136E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.112E-08 | 1.694E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.112E-08 | 1.27E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.774E-08 | 1.335E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.079E-08 | 1.235E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.949E-08 | 1.357E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 5.963E-08 | 1.793E-05 |
|---|
| base-excision repair | GO:0006284 |  | 1.827E-07 | 4.883E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.008E-06 | 2.424E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.719E-06 | 3.759E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.208E-08 | 2.952E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.283E-08 | 1.567E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.525E-08 | 1.242E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.525E-08 | 9.315E-06 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.004E-08 | 9.792E-06 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.224E-08 | 9.055E-06 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.853E-08 | 9.956E-06 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 4.308E-08 | 1.316E-05 |
|---|
| base-excision repair | GO:0006284 |  | 1.584E-07 | 4.3E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 8.866E-07 | 2.166E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.405E-06 | 3.121E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.673E-08 | 4.026E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.776E-08 | 2.136E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.112E-08 | 1.694E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.112E-08 | 1.27E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.774E-08 | 1.335E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.079E-08 | 1.235E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.949E-08 | 1.357E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 5.963E-08 | 1.793E-05 |
|---|
| base-excision repair | GO:0006284 |  | 1.827E-07 | 4.883E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.008E-06 | 2.424E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.719E-06 | 3.759E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.673E-08 | 4.026E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.776E-08 | 2.136E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.112E-08 | 1.694E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.112E-08 | 1.27E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.774E-08 | 1.335E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.079E-08 | 1.235E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.949E-08 | 1.357E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 5.963E-08 | 1.793E-05 |
|---|
| base-excision repair | GO:0006284 |  | 1.827E-07 | 4.883E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.008E-06 | 2.424E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.719E-06 | 3.759E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.346E-08 | 9.996E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.645E-08 | 5.341E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.632E-08 | 4.318E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.632E-08 | 3.238E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.182E-08 | 3.304E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.066E-08 | 3.092E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.008E-07 | 3.311E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.309E-07 | 3.764E-05 |
|---|
| base-excision repair | GO:0006284 |  | 3.724E-07 | 9.517E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.808E-06 | 4.158E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 7.938E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.346E-08 | 9.996E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.645E-08 | 5.341E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.632E-08 | 4.318E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.632E-08 | 3.238E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.182E-08 | 3.304E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.066E-08 | 3.092E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.008E-07 | 3.311E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.309E-07 | 3.764E-05 |
|---|
| base-excision repair | GO:0006284 |  | 3.724E-07 | 9.517E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.808E-06 | 4.158E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 7.938E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.346E-08 | 9.996E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.645E-08 | 5.341E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.632E-08 | 4.318E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.632E-08 | 3.238E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.182E-08 | 3.304E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.066E-08 | 3.092E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.008E-07 | 3.311E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.309E-07 | 3.764E-05 |
|---|
| base-excision repair | GO:0006284 |  | 3.724E-07 | 9.517E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.808E-06 | 4.158E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 7.938E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.346E-08 | 9.996E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.645E-08 | 5.341E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.632E-08 | 4.318E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.632E-08 | 3.238E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.182E-08 | 3.304E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.066E-08 | 3.092E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.008E-07 | 3.311E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.309E-07 | 3.764E-05 |
|---|
| base-excision repair | GO:0006284 |  | 3.724E-07 | 9.517E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.808E-06 | 4.158E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.796E-06 | 7.938E-04 |
|---|



