Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 54487-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1466 | 7.781e-03 | 2.293e-03 | 4.764e-02 |
|---|
| IPC-NIBC-129 | 0.1925 | 3.794e-02 | 2.920e-02 | 1.843e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2587 | 3.713e-02 | 2.831e-02 | 3.573e-01 |
|---|
| Loi_GPL570 | 0.2946 | 5.598e-02 | 7.357e-02 | 2.458e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1605 | 2.526e-02 | 1.684e-02 | 3.408e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2913 | 5.821e-02 | 2.719e-02 | 3.450e-01 |
|---|
| Schmidt | 0.2541 | 1.582e-02 | 1.700e-02 | 1.095e-01 |
|---|
| Sotiriou | 0.2991 | 1.199e-01 | 5.422e-02 | 6.273e-01 |
|---|
| Wang | 0.2201 | 3.374e-02 | 2.116e-02 | 1.328e-01 |
|---|
| Zhang | 0.3044 | 1.248e-02 | 6.520e-03 | 5.596e-02 |
|---|
Expression data for subnetwork 54487-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
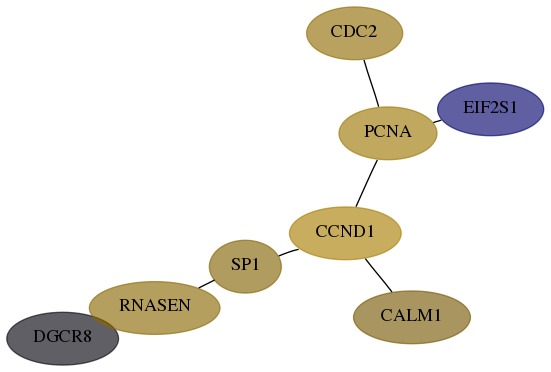
Score for each gene in subnetwork 54487-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| dgcr8 |   | 1 | 90 | 42 | 55 | -0.001 | 0.055 | 0.102 | -0.006 | 0.077 | -0.051 | 0.074 | 0.075 | 0.126 | 0.011 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| rnasen |   | 1 | 90 | 42 | 55 | 0.065 | 0.216 | 0.084 | -0.025 | 0.121 | 0.107 | 0.152 | 0.166 | 0.023 | 0.069 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
GO Enrichment output for subnetwork 54487-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.392E-07 | 1.24E-03 |
|---|
| gene silencing | GO:0016458 |  | 4.289E-06 | 4.933E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| response to dsRNA | GO:0043331 |  | 6.149E-05 | 0.02357018 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.04696989 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.633E-04 | 0.04693587 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.633E-04 | 0.04172077 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.078E-04 | 0.04780528 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.847E-04 | 0.05952061 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 1.828E-07 | 4.465E-04 |
|---|
| gene silencing | GO:0016458 |  | 1.324E-06 | 1.618E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.364E-06 | 3.554E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.585E-06 | 2.801E-03 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 6.543E-06 | 3.197E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.543E-06 | 2.664E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 4.259E-03 |
|---|
| response to dsRNA | GO:0043331 |  | 1.96E-05 | 5.985E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.213E-05 | 0.0141505 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.906E-05 | 0.01442756 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 8.24E-05 | 0.01830111 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 3.001E-07 | 7.221E-04 |
|---|
| gene silencing | GO:0016458 |  | 2.172E-06 | 2.613E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.077E-06 | 4.874E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.511E-06 | 4.518E-03 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 9.111E-06 | 4.384E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.111E-06 | 3.654E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 5.84E-03 |
|---|
| response to dsRNA | GO:0043331 |  | 2.728E-05 | 8.205E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.254E-05 | 0.01939114 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 8.217E-05 | 0.01976933 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.032E-04 | 0.0225753 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 1.828E-07 | 4.465E-04 |
|---|
| gene silencing | GO:0016458 |  | 1.324E-06 | 1.618E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.364E-06 | 3.554E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.585E-06 | 2.801E-03 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 6.543E-06 | 3.197E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.543E-06 | 2.664E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 4.259E-03 |
|---|
| response to dsRNA | GO:0043331 |  | 1.96E-05 | 5.985E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.213E-05 | 0.0141505 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 5.906E-05 | 0.01442756 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 8.24E-05 | 0.01830111 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 3.001E-07 | 7.221E-04 |
|---|
| gene silencing | GO:0016458 |  | 2.172E-06 | 2.613E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.077E-06 | 4.874E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.511E-06 | 4.518E-03 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 9.111E-06 | 4.384E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.111E-06 | 3.654E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 5.84E-03 |
|---|
| response to dsRNA | GO:0043331 |  | 2.728E-05 | 8.205E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.254E-05 | 0.01939114 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 8.217E-05 | 0.01976933 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.032E-04 | 0.0225753 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 3.001E-07 | 7.221E-04 |
|---|
| gene silencing | GO:0016458 |  | 2.172E-06 | 2.613E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.077E-06 | 4.874E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.511E-06 | 4.518E-03 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 9.111E-06 | 4.384E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.111E-06 | 3.654E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 5.84E-03 |
|---|
| response to dsRNA | GO:0043331 |  | 2.728E-05 | 8.205E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.254E-05 | 0.01939114 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 8.217E-05 | 0.01976933 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.032E-04 | 0.0225753 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.392E-07 | 1.24E-03 |
|---|
| gene silencing | GO:0016458 |  | 4.289E-06 | 4.933E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| response to dsRNA | GO:0043331 |  | 6.149E-05 | 0.02357018 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.04696989 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.633E-04 | 0.04693587 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.633E-04 | 0.04172077 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.078E-04 | 0.04780528 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.847E-04 | 0.05952061 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.392E-07 | 1.24E-03 |
|---|
| gene silencing | GO:0016458 |  | 4.289E-06 | 4.933E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| response to dsRNA | GO:0043331 |  | 6.149E-05 | 0.02357018 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.04696989 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.633E-04 | 0.04693587 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.633E-04 | 0.04172077 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.078E-04 | 0.04780528 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.847E-04 | 0.05952061 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.392E-07 | 1.24E-03 |
|---|
| gene silencing | GO:0016458 |  | 4.289E-06 | 4.933E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| response to dsRNA | GO:0043331 |  | 6.149E-05 | 0.02357018 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.04696989 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.633E-04 | 0.04693587 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.633E-04 | 0.04172077 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.078E-04 | 0.04780528 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.847E-04 | 0.05952061 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.392E-07 | 1.24E-03 |
|---|
| gene silencing | GO:0016458 |  | 4.289E-06 | 4.933E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| posttranscriptional gene silencing by RNA | GO:0035194 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| response to dsRNA | GO:0043331 |  | 6.149E-05 | 0.02357018 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.04696989 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.633E-04 | 0.04693587 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.633E-04 | 0.04172077 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.078E-04 | 0.04780528 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.847E-04 | 0.05952061 |
|---|



