Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5426-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1523 | 1.658e-02 | 6.440e-03 | 8.678e-02 |
|---|
| IPC-NIBC-129 | 0.2092 | 1.226e-01 | 1.107e-01 | 5.027e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2226 | 2.677e-02 | 1.932e-02 | 2.814e-01 |
|---|
| Loi_GPL570 | 0.2014 | 1.290e-01 | 1.583e-01 | 4.521e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1912 | 2.040e-02 | 1.270e-02 | 2.806e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3505 | 8.824e-02 | 4.637e-02 | 4.581e-01 |
|---|
| Schmidt | 0.1910 | 3.333e-02 | 3.519e-02 | 2.055e-01 |
|---|
| Sotiriou | 0.2520 | 7.331e-02 | 2.686e-02 | 4.707e-01 |
|---|
| Wang | 0.1852 | 6.161e-02 | 4.573e-02 | 2.111e-01 |
|---|
| Zhang | 0.2664 | 4.586e-03 | 2.132e-03 | 2.457e-02 |
|---|
Expression data for subnetwork 5426-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
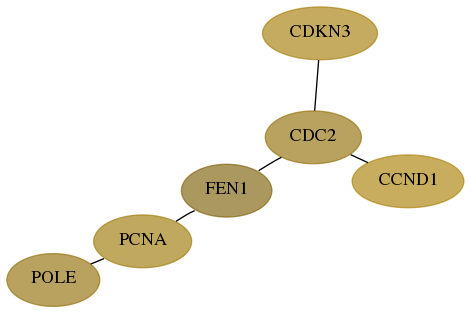
Score for each gene in subnetwork 5426-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| cdkn3 |   | 18 | 9 | 26 | 16 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.323 | 0.186 | 0.198 | 0.134 | 0.271 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| fen1 |   | 7 | 15 | 8 | 6 | 0.045 | 0.194 | 0.169 | 0.196 | 0.136 | 0.332 | 0.221 | 0.144 | 0.106 | 0.056 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| pole |   | 3 | 39 | 171 | 151 | 0.073 | 0.135 | 0.081 | -0.205 | 0.081 | 0.139 | 0.145 | 0.105 | 0.068 | -0.014 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 5426-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.59E-09 | 1.516E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.518E-08 | 8.646E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.121E-07 | 8.592E-05 |
|---|
| interphase | GO:0051325 |  | 1.23E-07 | 7.072E-05 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 9.291E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.767E-06 | 1.061E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.935E-06 | 9.644E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 1.775E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 1.929E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 1.893E-03 |
|---|
| UV protection | GO:0009650 |  | 1.322E-05 | 2.764E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.206E-09 | 2.945E-06 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.642E-08 | 2.006E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.644E-08 | 2.153E-05 |
|---|
| response to UV | GO:0009411 |  | 7.637E-07 | 4.664E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.514E-07 | 4.16E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 8.514E-07 | 3.467E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.958E-06 | 6.834E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 8.924E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.757E-06 | 1.02E-03 |
|---|
| UV protection | GO:0009650 |  | 5.737E-06 | 1.402E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.979E-05 | 4.394E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.932E-10 | 2.39E-06 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.473E-08 | 1.772E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.923E-08 | 1.542E-05 |
|---|
| interphase | GO:0051325 |  | 2.18E-08 | 1.312E-05 |
|---|
| response to UV | GO:0009411 |  | 6.121E-07 | 2.946E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.24E-07 | 2.903E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.641E-07 | 2.626E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.758E-06 | 5.286E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 7.42E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.568E-06 | 8.584E-04 |
|---|
| UV protection | GO:0009650 |  | 5.449E-06 | 1.192E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.206E-09 | 2.945E-06 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.642E-08 | 2.006E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.644E-08 | 2.153E-05 |
|---|
| response to UV | GO:0009411 |  | 7.637E-07 | 4.664E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.514E-07 | 4.16E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 8.514E-07 | 3.467E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.958E-06 | 6.834E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.922E-06 | 8.924E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.757E-06 | 1.02E-03 |
|---|
| UV protection | GO:0009650 |  | 5.737E-06 | 1.402E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.979E-05 | 4.394E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.932E-10 | 2.39E-06 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.473E-08 | 1.772E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.923E-08 | 1.542E-05 |
|---|
| interphase | GO:0051325 |  | 2.18E-08 | 1.312E-05 |
|---|
| response to UV | GO:0009411 |  | 6.121E-07 | 2.946E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.24E-07 | 2.903E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.641E-07 | 2.626E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.758E-06 | 5.286E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 7.42E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.568E-06 | 8.584E-04 |
|---|
| UV protection | GO:0009650 |  | 5.449E-06 | 1.192E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.932E-10 | 2.39E-06 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.473E-08 | 1.772E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.923E-08 | 1.542E-05 |
|---|
| interphase | GO:0051325 |  | 2.18E-08 | 1.312E-05 |
|---|
| response to UV | GO:0009411 |  | 6.121E-07 | 2.946E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.24E-07 | 2.903E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.641E-07 | 2.626E-04 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.758E-06 | 5.286E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 7.42E-04 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.568E-06 | 8.584E-04 |
|---|
| UV protection | GO:0009650 |  | 5.449E-06 | 1.192E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.59E-09 | 1.516E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.518E-08 | 8.646E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.121E-07 | 8.592E-05 |
|---|
| interphase | GO:0051325 |  | 1.23E-07 | 7.072E-05 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 9.291E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.767E-06 | 1.061E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.935E-06 | 9.644E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 1.775E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 1.929E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 1.893E-03 |
|---|
| UV protection | GO:0009650 |  | 1.322E-05 | 2.764E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.59E-09 | 1.516E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.518E-08 | 8.646E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.121E-07 | 8.592E-05 |
|---|
| interphase | GO:0051325 |  | 1.23E-07 | 7.072E-05 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 9.291E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.767E-06 | 1.061E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.935E-06 | 9.644E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 1.775E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 1.929E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 1.893E-03 |
|---|
| UV protection | GO:0009650 |  | 1.322E-05 | 2.764E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.59E-09 | 1.516E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.518E-08 | 8.646E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.121E-07 | 8.592E-05 |
|---|
| interphase | GO:0051325 |  | 1.23E-07 | 7.072E-05 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 9.291E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.767E-06 | 1.061E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.935E-06 | 9.644E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 1.775E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 1.929E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 1.893E-03 |
|---|
| UV protection | GO:0009650 |  | 1.322E-05 | 2.764E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.59E-09 | 1.516E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 7.518E-08 | 8.646E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.121E-07 | 8.592E-05 |
|---|
| interphase | GO:0051325 |  | 1.23E-07 | 7.072E-05 |
|---|
| response to UV | GO:0009411 |  | 2.02E-06 | 9.291E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.767E-06 | 1.061E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.935E-06 | 9.644E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 1.775E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 7.548E-06 | 1.929E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 8.231E-06 | 1.893E-03 |
|---|
| UV protection | GO:0009650 |  | 1.322E-05 | 2.764E-03 |
|---|



