Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 51-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1626 | 2.339e-02 | 1.027e-02 | 1.133e-01 |
|---|
| IPC-NIBC-129 | 0.2145 | 1.934e-01 | 1.842e-01 | 6.736e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2061 | 2.408e-02 | 1.707e-02 | 2.595e-01 |
|---|
| Loi_GPL570 | 0.1641 | 4.418e-02 | 5.917e-02 | 2.037e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1674 | 1.371e-02 | 7.496e-03 | 1.884e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3078 | 7.424e-02 | 3.715e-02 | 4.087e-01 |
|---|
| Schmidt | 0.2717 | 3.594e-02 | 3.789e-02 | 2.183e-01 |
|---|
| Sotiriou | 0.2472 | 1.075e-01 | 4.639e-02 | 5.914e-01 |
|---|
| Wang | 0.1933 | 5.368e-02 | 3.838e-02 | 1.902e-01 |
|---|
| Zhang | 0.2822 | 9.440e-03 | 4.774e-03 | 4.459e-02 |
|---|
Expression data for subnetwork 51-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
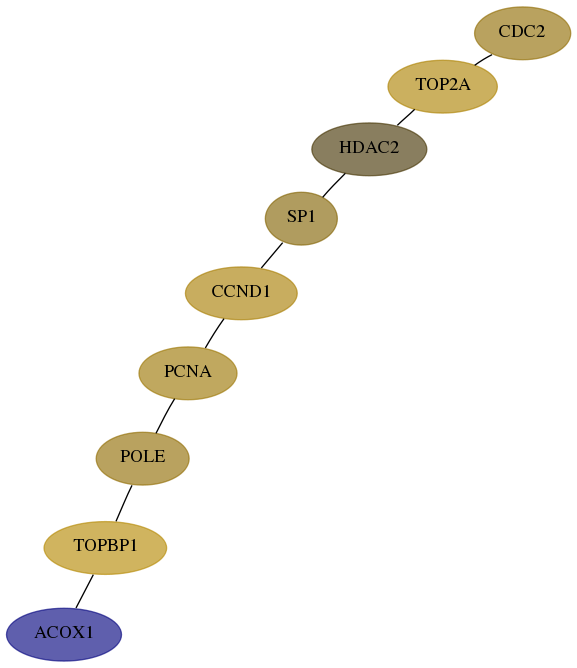
Score for each gene in subnetwork 51-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| pole |   | 3 | 39 | 171 | 151 | 0.073 | 0.135 | 0.081 | -0.205 | 0.081 | 0.139 | 0.145 | 0.105 | 0.068 | -0.014 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| hdac2 |   | 16 | 10 | 64 | 39 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 |
|---|
| acox1 |   | 1 | 90 | 180 | 186 | -0.046 | 0.072 | 0.062 | -0.053 | 0.100 | 0.010 | 0.194 | 0.140 | -0.036 | 0.169 |
|---|
| top2a |   | 22 | 7 | 70 | 46 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 |
|---|
| topbp1 |   | 1 | 90 | 180 | 186 | 0.173 | 0.189 | 0.113 | -0.073 | 0.106 | 0.115 | 0.178 | 0.093 | 0.117 | 0.091 |
|---|
GO Enrichment output for subnetwork 51-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.843E-07 | 6.538E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 7.105E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.861E-06 | 5.26E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.861E-06 | 3.945E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.097E-06 | 3.264E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.029E-05 | 3.943E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.101E-05 | 3.618E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.454E-05 | 3.717E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.918E-05 | 4.412E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.465E-05 | 5.155E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.491E-08 | 2.074E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.277E-06 | 2.781E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.593E-06 | 2.111E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.005E-06 | 1.835E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.005E-06 | 1.468E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.372E-06 | 1.78E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.506E-06 | 1.573E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.506E-06 | 1.376E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 7.132E-06 | 1.936E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 2.053E-03 |
|---|
| DNA topological change | GO:0006265 |  | 8.405E-06 | 1.867E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.327E-08 | 2.003E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-06 | 2.687E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.543E-06 | 2.04E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.996E-06 | 1.802E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.996E-06 | 1.442E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.996E-06 | 1.201E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.065E-06 | 1.397E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.492E-06 | 1.351E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 6.998E-06 | 1.871E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.38E-06 | 2.016E-03 |
|---|
| DNA topological change | GO:0006265 |  | 8.38E-06 | 1.833E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.491E-08 | 2.074E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.277E-06 | 2.781E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.593E-06 | 2.111E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.005E-06 | 1.835E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.005E-06 | 1.468E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.372E-06 | 1.78E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.506E-06 | 1.573E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.506E-06 | 1.376E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 7.132E-06 | 1.936E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 2.053E-03 |
|---|
| DNA topological change | GO:0006265 |  | 8.405E-06 | 1.867E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.327E-08 | 2.003E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-06 | 2.687E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.543E-06 | 2.04E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.996E-06 | 1.802E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.996E-06 | 1.442E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.996E-06 | 1.201E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.065E-06 | 1.397E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.492E-06 | 1.351E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 6.998E-06 | 1.871E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.38E-06 | 2.016E-03 |
|---|
| DNA topological change | GO:0006265 |  | 8.38E-06 | 1.833E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.327E-08 | 2.003E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-06 | 2.687E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.543E-06 | 2.04E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.996E-06 | 1.802E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.996E-06 | 1.442E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.996E-06 | 1.201E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.065E-06 | 1.397E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.492E-06 | 1.351E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 6.998E-06 | 1.871E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.38E-06 | 2.016E-03 |
|---|
| DNA topological change | GO:0006265 |  | 8.38E-06 | 1.833E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.843E-07 | 6.538E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 7.105E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.861E-06 | 5.26E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.861E-06 | 3.945E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.097E-06 | 3.264E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.029E-05 | 3.943E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.101E-05 | 3.618E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.454E-05 | 3.717E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.918E-05 | 4.412E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.465E-05 | 5.155E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.843E-07 | 6.538E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 7.105E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.861E-06 | 5.26E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.861E-06 | 3.945E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.097E-06 | 3.264E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.029E-05 | 3.943E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.101E-05 | 3.618E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.454E-05 | 3.717E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.918E-05 | 4.412E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.465E-05 | 5.155E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.843E-07 | 6.538E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 7.105E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.861E-06 | 5.26E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.861E-06 | 3.945E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.097E-06 | 3.264E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.029E-05 | 3.943E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.101E-05 | 3.618E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.454E-05 | 3.717E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.918E-05 | 4.412E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.465E-05 | 5.155E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.843E-07 | 6.538E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 7.105E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.861E-06 | 5.26E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.861E-06 | 3.945E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.097E-06 | 3.264E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.029E-05 | 3.943E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.101E-05 | 3.618E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.454E-05 | 3.717E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.918E-05 | 4.412E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.465E-05 | 5.155E-03 |
|---|



