Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5036-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1174 | 1.854e-02 | 7.498e-03 | 9.469e-02 |
|---|
| IPC-NIBC-129 | 0.1989 | 5.067e-02 | 4.064e-02 | 2.425e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2172 | 3.274e-02 | 2.445e-02 | 3.267e-01 |
|---|
| Loi_GPL570 | 0.2719 | 7.411e-02 | 9.520e-02 | 3.048e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1795 | 7.357e-02 | 6.693e-02 | 7.104e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3071 | 8.643e-02 | 4.515e-02 | 4.520e-01 |
|---|
| Schmidt | 0.2331 | 3.775e-02 | 3.975e-02 | 2.270e-01 |
|---|
| Sotiriou | 0.2441 | 8.849e-02 | 3.515e-02 | 5.288e-01 |
|---|
| Wang | 0.1950 | 5.208e-02 | 3.693e-02 | 1.859e-01 |
|---|
| Zhang | 0.2683 | 9.553e-03 | 4.837e-03 | 4.502e-02 |
|---|
Expression data for subnetwork 5036-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
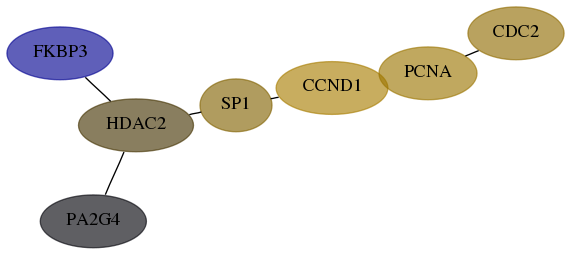
Score for each gene in subnetwork 5036-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| hdac2 |   | 16 | 10 | 64 | 39 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| pa2g4 |   | 1 | 90 | 163 | 172 | -0.001 | 0.092 | 0.146 | 0.101 | 0.134 | 0.212 | 0.129 | 0.108 | 0.032 | -0.012 |
|---|
| fkbp3 |   | 2 | 53 | 129 | 116 | -0.072 | 0.129 | 0.144 | 0.166 | 0.136 | 0.010 | 0.159 | 0.052 | 0.058 | 0.199 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 5036-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 0.01041131 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.364E-05 | 0.0181276 |
|---|
| histone deacetylation | GO:0016575 |  | 2.367E-05 | 0.01361307 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.449E-05 | 0.01126533 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01475281 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.513E-05 | 0.01482798 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.156E-05 | 0.01482233 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.156E-05 | 0.0131754 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.156E-05 | 0.01185786 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 6.568E-05 | 0.01373386 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.547E-07 | 1.6E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.234E-06 | 1.507E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.851E-06 | 1.507E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.453E-06 | 2.109E-03 |
|---|
| histone deacetylation | GO:0016575 |  | 1.293E-05 | 6.318E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.478E-05 | 6.016E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.674E-05 | 5.843E-03 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 2.337E-05 | 7.138E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.337E-05 | 6.345E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.841E-05 | 6.94E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.993E-05 | 8.869E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.074E-06 | 2.585E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.719E-06 | 2.068E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.577E-06 | 2.067E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 2.893E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.916E-06 | 4.772E-03 |
|---|
| histone deacetylation | GO:0016575 |  | 1.8E-05 | 7.219E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.057E-05 | 7.07E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.331E-05 | 7.009E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.929E-05 | 7.83E-03 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 3.254E-05 | 7.828E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 3.954E-05 | 8.648E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.547E-07 | 1.6E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.234E-06 | 1.507E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.851E-06 | 1.507E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.453E-06 | 2.109E-03 |
|---|
| histone deacetylation | GO:0016575 |  | 1.293E-05 | 6.318E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.478E-05 | 6.016E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.674E-05 | 5.843E-03 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 2.337E-05 | 7.138E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.337E-05 | 6.345E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.841E-05 | 6.94E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.993E-05 | 8.869E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.074E-06 | 2.585E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.719E-06 | 2.068E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.577E-06 | 2.067E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 2.893E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.916E-06 | 4.772E-03 |
|---|
| histone deacetylation | GO:0016575 |  | 1.8E-05 | 7.219E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.057E-05 | 7.07E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.331E-05 | 7.009E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.929E-05 | 7.83E-03 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 3.254E-05 | 7.828E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 3.954E-05 | 8.648E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.074E-06 | 2.585E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.719E-06 | 2.068E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.577E-06 | 2.067E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.809E-06 | 2.893E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.916E-06 | 4.772E-03 |
|---|
| histone deacetylation | GO:0016575 |  | 1.8E-05 | 7.219E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.057E-05 | 7.07E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 2.331E-05 | 7.009E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.929E-05 | 7.83E-03 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 3.254E-05 | 7.828E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 3.954E-05 | 8.648E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 0.01041131 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.364E-05 | 0.0181276 |
|---|
| histone deacetylation | GO:0016575 |  | 2.367E-05 | 0.01361307 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.449E-05 | 0.01126533 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01475281 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.513E-05 | 0.01482798 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.156E-05 | 0.01482233 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.156E-05 | 0.0131754 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.156E-05 | 0.01185786 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 6.568E-05 | 0.01373386 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 0.01041131 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.364E-05 | 0.0181276 |
|---|
| histone deacetylation | GO:0016575 |  | 2.367E-05 | 0.01361307 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.449E-05 | 0.01126533 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01475281 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.513E-05 | 0.01482798 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.156E-05 | 0.01482233 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.156E-05 | 0.0131754 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.156E-05 | 0.01185786 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 6.568E-05 | 0.01373386 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 0.01041131 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.364E-05 | 0.0181276 |
|---|
| histone deacetylation | GO:0016575 |  | 2.367E-05 | 0.01361307 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.449E-05 | 0.01126533 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01475281 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.513E-05 | 0.01482798 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.156E-05 | 0.01482233 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.156E-05 | 0.0131754 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.156E-05 | 0.01185786 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 6.568E-05 | 0.01373386 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.45E-06 | 7.935E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.053E-06 | 0.01041131 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.364E-05 | 0.0181276 |
|---|
| histone deacetylation | GO:0016575 |  | 2.367E-05 | 0.01361307 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.449E-05 | 0.01126533 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01475281 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.513E-05 | 0.01482798 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.156E-05 | 0.01482233 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.156E-05 | 0.0131754 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.156E-05 | 0.01185786 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 6.568E-05 | 0.01373386 |
|---|



