Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3178-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1535 | 1.244e-02 | 4.357e-03 | 6.927e-02 |
|---|
| IPC-NIBC-129 | 0.2223 | 8.621e-02 | 7.434e-02 | 3.844e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2364 | 2.065e-02 | 1.427e-02 | 2.300e-01 |
|---|
| Loi_GPL570 | 0.2298 | 3.318e-02 | 4.546e-02 | 1.610e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1501 | 1.945e-02 | 1.192e-02 | 2.681e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3319 | 6.654e-02 | 3.229e-02 | 3.792e-01 |
|---|
| Schmidt | 0.2928 | 3.313e-02 | 3.498e-02 | 2.045e-01 |
|---|
| Sotiriou | 0.2524 | 1.412e-01 | 6.842e-02 | 6.810e-01 |
|---|
| Wang | 0.2300 | 2.840e-02 | 1.695e-02 | 1.160e-01 |
|---|
| Zhang | 0.2922 | 6.288e-03 | 3.032e-03 | 3.192e-02 |
|---|
Expression data for subnetwork 3178-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
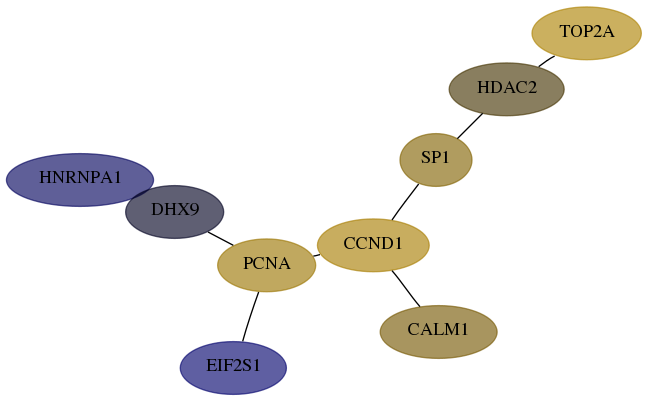
Score for each gene in subnetwork 3178-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| dhx9 |   | 1 | 90 | 70 | 82 | -0.004 | 0.165 | 0.099 | -0.094 | 0.103 | 0.127 | 0.155 | 0.135 | 0.075 | 0.007 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| hnrnpa1 |   | 2 | 53 | 70 | 60 | -0.021 | 0.209 | 0.088 | -0.020 | 0.087 | 0.082 | 0.036 | 0.089 | 0.077 | 0.083 |
|---|
| hdac2 |   | 16 | 10 | 64 | 39 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 |
|---|
| top2a |   | 22 | 7 | 70 | 46 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
GO Enrichment output for subnetwork 3178-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 0.03154099 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 0.0157705 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| DNA topological change | GO:0006265 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| chromosome segregation | GO:0007059 |  | 4.192E-05 | 0.01607081 |
|---|
| DNA ligation | GO:0006266 |  | 4.923E-05 | 0.01617412 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 6.149E-05 | 0.01767763 |
|---|
| histone deacetylation | GO:0016575 |  | 7.51E-05 | 0.01919142 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 8.101E-05 | 0.01863257 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.02988993 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.743E-06 | 0.01158756 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.743E-06 | 5.794E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.206E-06 | 4.239E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.112E-06 | 4.343E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 7.112E-06 | 3.475E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 5.401E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.326E-05 | 4.629E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.429E-05 | 4.364E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 2.007E-05 | 5.448E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.13E-05 | 5.203E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.13E-05 | 4.73E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.605E-06 | 0.01589185 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.605E-06 | 7.946E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.605E-06 | 5.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 5.128E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.903E-06 | 4.765E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 7.405E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.847E-05 | 6.347E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.337E-05 | 7.028E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.965E-05 | 7.926E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.965E-05 | 7.133E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.28E-05 | 7.173E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.743E-06 | 0.01158756 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.743E-06 | 5.794E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.206E-06 | 4.239E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.112E-06 | 4.343E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 7.112E-06 | 3.475E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 5.401E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.326E-05 | 4.629E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.429E-05 | 4.364E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 2.007E-05 | 5.448E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.13E-05 | 5.203E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.13E-05 | 4.73E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.605E-06 | 0.01589185 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.605E-06 | 7.946E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.605E-06 | 5.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 5.128E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.903E-06 | 4.765E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 7.405E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.847E-05 | 6.347E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.337E-05 | 7.028E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.965E-05 | 7.926E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.965E-05 | 7.133E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.28E-05 | 7.173E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.605E-06 | 0.01589185 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.605E-06 | 7.946E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.605E-06 | 5.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 5.128E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.903E-06 | 4.765E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 7.405E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.847E-05 | 6.347E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.337E-05 | 7.028E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.965E-05 | 7.926E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.965E-05 | 7.133E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.28E-05 | 7.173E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 0.03154099 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 0.0157705 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| DNA topological change | GO:0006265 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| chromosome segregation | GO:0007059 |  | 4.192E-05 | 0.01607081 |
|---|
| DNA ligation | GO:0006266 |  | 4.923E-05 | 0.01617412 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 6.149E-05 | 0.01767763 |
|---|
| histone deacetylation | GO:0016575 |  | 7.51E-05 | 0.01919142 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 8.101E-05 | 0.01863257 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.02988993 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 0.03154099 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 0.0157705 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| DNA topological change | GO:0006265 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| chromosome segregation | GO:0007059 |  | 4.192E-05 | 0.01607081 |
|---|
| DNA ligation | GO:0006266 |  | 4.923E-05 | 0.01617412 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 6.149E-05 | 0.01767763 |
|---|
| histone deacetylation | GO:0016575 |  | 7.51E-05 | 0.01919142 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 8.101E-05 | 0.01863257 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.02988993 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 0.03154099 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 0.0157705 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| DNA topological change | GO:0006265 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| chromosome segregation | GO:0007059 |  | 4.192E-05 | 0.01607081 |
|---|
| DNA ligation | GO:0006266 |  | 4.923E-05 | 0.01617412 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 6.149E-05 | 0.01767763 |
|---|
| histone deacetylation | GO:0016575 |  | 7.51E-05 | 0.01919142 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 8.101E-05 | 0.01863257 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.02988993 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 0.03154099 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 0.0157705 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.052E-05 | 0.01573263 |
|---|
| DNA topological change | GO:0006265 |  | 2.056E-05 | 0.0118193 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01322803 |
|---|
| chromosome segregation | GO:0007059 |  | 4.192E-05 | 0.01607081 |
|---|
| DNA ligation | GO:0006266 |  | 4.923E-05 | 0.01617412 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 6.149E-05 | 0.01767763 |
|---|
| histone deacetylation | GO:0016575 |  | 7.51E-05 | 0.01919142 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 8.101E-05 | 0.01863257 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.43E-04 | 0.02988993 |
|---|



