Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3159-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1878 | 1.108e-02 | 3.718e-03 | 6.319e-02 |
|---|
| IPC-NIBC-129 | 0.1508 | 1.068e-01 | 9.470e-02 | 4.543e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2419 | 8.344e-02 | 7.229e-02 | 5.904e-01 |
|---|
| Loi_GPL570 | 0.2126 | 8.244e-02 | 1.050e-01 | 3.299e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1911 | 5.059e-03 | 1.960e-03 | 5.772e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3315 | 1.352e-01 | 8.009e-02 | 5.919e-01 |
|---|
| Schmidt | 0.2251 | 2.480e-02 | 2.637e-02 | 1.611e-01 |
|---|
| Sotiriou | 0.2708 | 7.344e-02 | 2.693e-02 | 4.712e-01 |
|---|
| Wang | 0.1399 | 1.329e-01 | 1.198e-01 | 3.686e-01 |
|---|
| Zhang | 0.2268 | 6.327e-03 | 3.053e-03 | 3.209e-02 |
|---|
Expression data for subnetwork 3159-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
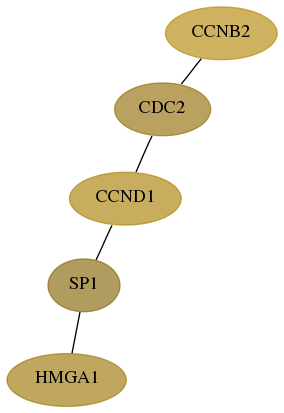
Score for each gene in subnetwork 3159-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| hmga1 |   | 1 | 90 | 208 | 208 | 0.092 | 0.071 | 0.202 | 0.017 | 0.101 | 0.159 | 0.151 | 0.107 | -0.054 | -0.051 |
|---|
| ccnb2 |   | 4 | 27 | 188 | 164 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 3159-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.08E-07 | 7.083E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 4.412E-06 | 3.383E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 4.412E-06 | 2.537E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.841E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 7.873E-06 | 3.018E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.231E-06 | 2.704E-03 |
|---|
| thymus development | GO:0048538 |  | 1.058E-05 | 3.042E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.615E-05 | 4.128E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.615E-05 | 3.715E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.289E-05 | 4.787E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 6.423E-08 | 1.569E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.785E-07 | 4.623E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.703E-07 | 7.087E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.305E-06 | 7.971E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.788E-06 | 8.737E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.827E-06 | 7.439E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 8.5E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.436E-06 | 7.437E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.131E-06 | 8.499E-04 |
|---|
| thymus development | GO:0048538 |  | 4.782E-06 | 1.168E-03 |
|---|
| lysogeny | GO:0030069 |  | 4.782E-06 | 1.062E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 7.678E-08 | 1.847E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.524E-07 | 5.442E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 9.918E-07 | 7.954E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.487E-06 | 8.946E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.979E-06 | 9.524E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 2.082E-06 | 8.349E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 9.54E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.775E-06 | 8.347E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.568E-06 | 9.538E-04 |
|---|
| thymus development | GO:0048538 |  | 5.449E-06 | 1.311E-03 |
|---|
| lysogeny | GO:0030069 |  | 5.449E-06 | 1.192E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 6.423E-08 | 1.569E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.785E-07 | 4.623E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.703E-07 | 7.087E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.305E-06 | 7.971E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.788E-06 | 8.737E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 1.827E-06 | 7.439E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 8.5E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.436E-06 | 7.437E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.131E-06 | 8.499E-04 |
|---|
| thymus development | GO:0048538 |  | 4.782E-06 | 1.168E-03 |
|---|
| lysogeny | GO:0030069 |  | 4.782E-06 | 1.062E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 7.678E-08 | 1.847E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.524E-07 | 5.442E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 9.918E-07 | 7.954E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.487E-06 | 8.946E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.979E-06 | 9.524E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 2.082E-06 | 8.349E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 9.54E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.775E-06 | 8.347E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.568E-06 | 9.538E-04 |
|---|
| thymus development | GO:0048538 |  | 5.449E-06 | 1.311E-03 |
|---|
| lysogeny | GO:0030069 |  | 5.449E-06 | 1.192E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 7.678E-08 | 1.847E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.524E-07 | 5.442E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 9.918E-07 | 7.954E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.487E-06 | 8.946E-04 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 1.979E-06 | 9.524E-04 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 2.082E-06 | 8.349E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 9.54E-04 |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.775E-06 | 8.347E-04 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 3.568E-06 | 9.538E-04 |
|---|
| thymus development | GO:0048538 |  | 5.449E-06 | 1.311E-03 |
|---|
| lysogeny | GO:0030069 |  | 5.449E-06 | 1.192E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.08E-07 | 7.083E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 4.412E-06 | 3.383E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 4.412E-06 | 2.537E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.841E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 7.873E-06 | 3.018E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.231E-06 | 2.704E-03 |
|---|
| thymus development | GO:0048538 |  | 1.058E-05 | 3.042E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.615E-05 | 4.128E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.615E-05 | 3.715E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.289E-05 | 4.787E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.08E-07 | 7.083E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 4.412E-06 | 3.383E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 4.412E-06 | 2.537E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.841E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 7.873E-06 | 3.018E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.231E-06 | 2.704E-03 |
|---|
| thymus development | GO:0048538 |  | 1.058E-05 | 3.042E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.615E-05 | 4.128E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.615E-05 | 3.715E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.289E-05 | 4.787E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.08E-07 | 7.083E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 4.412E-06 | 3.383E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 4.412E-06 | 2.537E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.841E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 7.873E-06 | 3.018E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.231E-06 | 2.704E-03 |
|---|
| thymus development | GO:0048538 |  | 1.058E-05 | 3.042E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.615E-05 | 4.128E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.615E-05 | 3.715E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.289E-05 | 4.787E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of chromosome organization | GO:0033044 |  | 3.08E-07 | 7.083E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 2.172E-03 |
|---|
| positive regulation of gene expression. epigenetic | GO:0045815 |  | 4.412E-06 | 3.383E-03 |
|---|
| regulation of chromatin assembly or disassembly | GO:0001672 |  | 4.412E-06 | 2.537E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 2.841E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 7.873E-06 | 3.018E-03 |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.231E-06 | 2.704E-03 |
|---|
| thymus development | GO:0048538 |  | 1.058E-05 | 3.042E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.615E-05 | 4.128E-03 |
|---|
| lysogeny | GO:0030069 |  | 1.615E-05 | 3.715E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.289E-05 | 4.787E-03 |
|---|



