Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 29984-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1600 | 1.046e-02 | 3.439e-03 | 6.037e-02 |
|---|
| IPC-NIBC-129 | 0.2318 | 1.089e-01 | 9.676e-02 | 4.608e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2446 | 1.708e-02 | 1.142e-02 | 1.972e-01 |
|---|
| Loi_GPL570 | 0.2111 | 2.318e-02 | 3.266e-02 | 1.187e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1265 | 1.518e-02 | 8.585e-03 | 2.095e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2544 | 2.522e-01 | 1.780e-01 | 7.960e-01 |
|---|
| Schmidt | 0.3191 | 2.656e-02 | 2.819e-02 | 1.706e-01 |
|---|
| Sotiriou | 0.2664 | 2.040e-01 | 1.154e-01 | 7.973e-01 |
|---|
| Wang | 0.2144 | 3.728e-02 | 2.406e-02 | 1.436e-01 |
|---|
| Zhang | 0.1672 | 2.314e-02 | 1.303e-02 | 9.188e-02 |
|---|
Expression data for subnetwork 29984-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
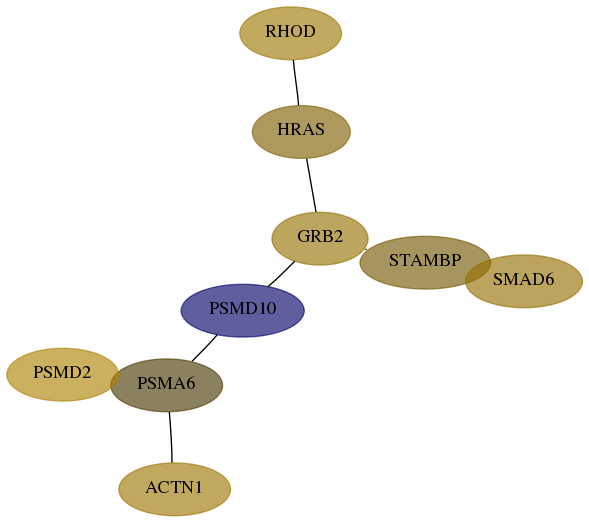
Score for each gene in subnetwork 29984-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| psma6 |   | 2 | 53 | 138 | 125 | 0.013 | 0.229 | 0.165 | 0.171 | 0.108 | 0.235 | 0.133 | 0.047 | 0.127 | 0.187 |
|---|
| psmd2 |   | 3 | 39 | 20 | 22 | 0.147 | 0.322 | 0.122 | 0.146 | 0.143 | 0.194 | 0.318 | 0.151 | 0.154 | 0.042 |
|---|
| psmd10 |   | 2 | 53 | 32 | 35 | -0.028 | 0.099 | 0.108 | 0.130 | 0.137 | 0.153 | 0.061 | 0.191 | 0.075 | 0.135 |
|---|
| rhod |   | 1 | 90 | 198 | 198 | 0.102 | -0.027 | 0.115 | 0.119 | 0.082 | 0.043 | 0.037 | 0.082 | 0.093 | 0.008 |
|---|
| smad6 |   | 2 | 53 | 145 | 130 | 0.083 | 0.077 | 0.039 | -0.005 | 0.051 | -0.105 | 0.121 | -0.019 | 0.161 | 0.065 |
|---|
| hras |   | 5 | 20 | 51 | 36 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.245 | 0.086 | 0.208 | 0.001 | 0.003 |
|---|
| actn1 |   | 1 | 90 | 198 | 198 | 0.100 | 0.223 | 0.031 | -0.052 | 0.053 | 0.019 | 0.024 | 0.027 | 0.124 | -0.008 |
|---|
| stambp |   | 2 | 53 | 145 | 130 | 0.038 | 0.150 | 0.160 | 0.131 | 0.100 | 0.223 | 0.284 | 0.065 | 0.076 | 0.050 |
|---|
| grb2 |   | 6 | 17 | 8 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 |
|---|
GO Enrichment output for subnetwork 29984-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 6.078E-08 | 1.398E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.502E-08 | 7.478E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.502E-08 | 4.985E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.948E-08 | 3.995E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.424E-08 | 3.875E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.424E-08 | 3.229E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.074E-07 | 3.529E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.206E-07 | 3.468E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.507E-07 | 3.851E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.957E-07 | 4.501E-05 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 4.816E-07 | 1.007E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.063E-08 | 5.039E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.191E-08 | 2.676E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.191E-08 | 1.784E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.325E-08 | 1.42E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.765E-08 | 1.351E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.765E-08 | 1.126E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.632E-08 | 1.267E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.03E-08 | 1.231E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.168E-08 | 1.403E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 7.802E-08 | 1.906E-05 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 5.311E-06 | 1.179E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.997E-08 | 7.211E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 3.183E-08 | 3.83E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.183E-08 | 2.553E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.378E-08 | 2.032E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.017E-08 | 1.933E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.017E-08 | 1.611E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 5.276E-08 | 1.813E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 5.854E-08 | 1.761E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 7.507E-08 | 2.007E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.133E-07 | 2.726E-05 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.472E-06 | 1.416E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.063E-08 | 5.039E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.191E-08 | 2.676E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.191E-08 | 1.784E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.325E-08 | 1.42E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.765E-08 | 1.351E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.765E-08 | 1.126E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.632E-08 | 1.267E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.03E-08 | 1.231E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.168E-08 | 1.403E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 7.802E-08 | 1.906E-05 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 5.311E-06 | 1.179E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.997E-08 | 7.211E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 3.183E-08 | 3.83E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.183E-08 | 2.553E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.378E-08 | 2.032E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.017E-08 | 1.933E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.017E-08 | 1.611E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 5.276E-08 | 1.813E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 5.854E-08 | 1.761E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 7.507E-08 | 2.007E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.133E-07 | 2.726E-05 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.472E-06 | 1.416E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.997E-08 | 7.211E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 3.183E-08 | 3.83E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.183E-08 | 2.553E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.378E-08 | 2.032E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.017E-08 | 1.933E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.017E-08 | 1.611E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 5.276E-08 | 1.813E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 5.854E-08 | 1.761E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 7.507E-08 | 2.007E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.133E-07 | 2.726E-05 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.472E-06 | 1.416E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 6.078E-08 | 1.398E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.502E-08 | 7.478E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.502E-08 | 4.985E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.948E-08 | 3.995E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.424E-08 | 3.875E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.424E-08 | 3.229E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.074E-07 | 3.529E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.206E-07 | 3.468E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.507E-07 | 3.851E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.957E-07 | 4.501E-05 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 4.816E-07 | 1.007E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 6.078E-08 | 1.398E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.502E-08 | 7.478E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.502E-08 | 4.985E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.948E-08 | 3.995E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.424E-08 | 3.875E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.424E-08 | 3.229E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.074E-07 | 3.529E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.206E-07 | 3.468E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.507E-07 | 3.851E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.957E-07 | 4.501E-05 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 4.816E-07 | 1.007E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 6.078E-08 | 1.398E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.502E-08 | 7.478E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.502E-08 | 4.985E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.948E-08 | 3.995E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.424E-08 | 3.875E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.424E-08 | 3.229E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.074E-07 | 3.529E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.206E-07 | 3.468E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.507E-07 | 3.851E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.957E-07 | 4.501E-05 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 4.816E-07 | 1.007E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 6.078E-08 | 1.398E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.502E-08 | 7.478E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.502E-08 | 4.985E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.948E-08 | 3.995E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.424E-08 | 3.875E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.424E-08 | 3.229E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.074E-07 | 3.529E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.206E-07 | 3.468E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.507E-07 | 3.851E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.957E-07 | 4.501E-05 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 4.816E-07 | 1.007E-04 |
|---|



