Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 29966-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2062 | 1.079e-02 | 3.587e-03 | 6.188e-02 |
|---|
| IPC-NIBC-129 | 0.2100 | 3.348e-02 | 2.532e-02 | 1.630e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2432 | 2.635e-02 | 1.897e-02 | 2.780e-01 |
|---|
| Loi_GPL570 | 0.3043 | 1.456e-01 | 1.768e-01 | 4.894e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1408 | 2.403e-03 | 7.110e-04 | 2.072e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.2396 | 1.644e-01 | 1.029e-01 | 6.567e-01 |
|---|
| Schmidt | 0.1816 | 8.480e-03 | 9.241e-03 | 6.228e-02 |
|---|
| Sotiriou | 0.3380 | 1.633e-01 | 8.417e-02 | 7.282e-01 |
|---|
| Wang | 0.1936 | 5.333e-02 | 3.806e-02 | 1.893e-01 |
|---|
| Zhang | 0.2084 | 2.958e-02 | 1.715e-02 | 1.115e-01 |
|---|
Expression data for subnetwork 29966-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
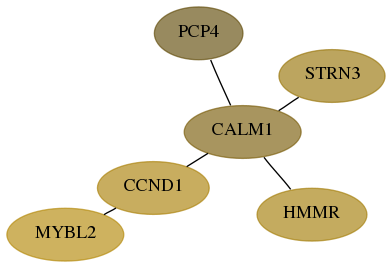
Score for each gene in subnetwork 29966-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| pcp4 |   | 5 | 20 | 35 | 25 | 0.022 | 0.163 | 0.112 | 0.140 | 0.114 | -0.002 | 0.096 | 0.117 | 0.104 | 0.109 |
|---|
| mybl2 |   | 5 | 20 | 92 | 63 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.275 | 0.202 | 0.182 | 0.120 | 0.074 |
|---|
| strn3 |   | 1 | 90 | 132 | 137 | 0.084 | 0.042 | 0.062 | 0.267 | 0.076 | -0.048 | -0.010 | 0.103 | 0.167 | 0.030 |
|---|
| hmmr |   | 10 | 14 | 35 | 23 | 0.112 | 0.310 | 0.147 | 0.127 | 0.145 | 0.221 | 0.179 | 0.233 | 0.227 | 0.144 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 29966-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.08166013 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.09505239 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.08580805 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.07485276 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.17499778 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.17341346 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.1662038 |
|---|
| response to UV | GO:0009411 |  | 6.041E-04 | 0.15438137 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.425E-04 | 0.1707709 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.167E-03 | 0.24401646 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 0.01653866 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.577E-05 | 0.05591161 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.815E-05 | 0.06364417 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.085E-05 | 0.05548399 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.755E-05 | 0.04766083 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.474E-04 | 0.10073489 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.695E-04 | 0.09404622 |
|---|
| response to calcium ion | GO:0051592 |  | 2.809E-04 | 0.08576644 |
|---|
| response to UV | GO:0009411 |  | 3.54E-04 | 0.09609537 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.802E-04 | 0.0928947 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 6.402E-04 | 0.14219233 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.05414716 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.06321238 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.05543449 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.04774415 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.702E-04 | 0.10833418 |
|---|
| response to calcium ion | GO:0051592 |  | 2.821E-04 | 0.09695485 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.943E-04 | 0.08849657 |
|---|
| response to UV | GO:0009411 |  | 3.589E-04 | 0.09594921 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.152E-04 | 0.09989173 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 6.618E-04 | 0.14475956 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 0.01653866 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.577E-05 | 0.05591161 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.815E-05 | 0.06364417 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.085E-05 | 0.05548399 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.755E-05 | 0.04766083 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.474E-04 | 0.10073489 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.695E-04 | 0.09404622 |
|---|
| response to calcium ion | GO:0051592 |  | 2.809E-04 | 0.08576644 |
|---|
| response to UV | GO:0009411 |  | 3.54E-04 | 0.09609537 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.802E-04 | 0.0928947 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 6.402E-04 | 0.14219233 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.05414716 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.06321238 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.05543449 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.04774415 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.702E-04 | 0.10833418 |
|---|
| response to calcium ion | GO:0051592 |  | 2.821E-04 | 0.09695485 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.943E-04 | 0.08849657 |
|---|
| response to UV | GO:0009411 |  | 3.589E-04 | 0.09594921 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.152E-04 | 0.09989173 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 6.618E-04 | 0.14475956 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.05414716 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.06321238 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.05543449 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.04774415 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.702E-04 | 0.10833418 |
|---|
| response to calcium ion | GO:0051592 |  | 2.821E-04 | 0.09695485 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.943E-04 | 0.08849657 |
|---|
| response to UV | GO:0009411 |  | 3.589E-04 | 0.09594921 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.152E-04 | 0.09989173 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 6.618E-04 | 0.14475956 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.08166013 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.09505239 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.08580805 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.07485276 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.17499778 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.17341346 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.1662038 |
|---|
| response to UV | GO:0009411 |  | 6.041E-04 | 0.15438137 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.425E-04 | 0.1707709 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.167E-03 | 0.24401646 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.08166013 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.09505239 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.08580805 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.07485276 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.17499778 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.17341346 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.1662038 |
|---|
| response to UV | GO:0009411 |  | 6.041E-04 | 0.15438137 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.425E-04 | 0.1707709 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.167E-03 | 0.24401646 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.08166013 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.09505239 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.08580805 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.07485276 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.17499778 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.17341346 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.1662038 |
|---|
| response to UV | GO:0009411 |  | 6.041E-04 | 0.15438137 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.425E-04 | 0.1707709 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.167E-03 | 0.24401646 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.08166013 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.09505239 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.08580805 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.07485276 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.17499778 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.17341346 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.1662038 |
|---|
| response to UV | GO:0009411 |  | 6.041E-04 | 0.15438137 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.425E-04 | 0.1707709 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.167E-03 | 0.24401646 |
|---|



