Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2968-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1636 | 1.510e-02 | 5.669e-03 | 8.065e-02 |
|---|
| IPC-NIBC-129 | 0.1857 | 1.312e-01 | 1.194e-01 | 5.272e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2271 | 4.248e-02 | 3.311e-02 | 3.920e-01 |
|---|
| Loi_GPL570 | 0.1959 | 8.534e-02 | 1.084e-01 | 3.383e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2075 | 1.321e-02 | 7.131e-03 | 1.810e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3532 | 1.688e-01 | 1.065e-01 | 6.656e-01 |
|---|
| Schmidt | 0.2225 | 2.850e-02 | 3.020e-02 | 1.809e-01 |
|---|
| Sotiriou | 0.2620 | 5.633e-02 | 1.843e-02 | 3.948e-01 |
|---|
| Wang | 0.1803 | 6.708e-02 | 5.094e-02 | 2.250e-01 |
|---|
| Zhang | 0.2059 | 4.376e-03 | 2.024e-03 | 2.363e-02 |
|---|
Expression data for subnetwork 2968-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
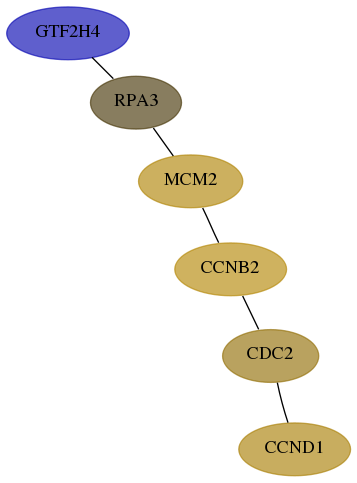
Score for each gene in subnetwork 2968-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| ccnb2 |   | 4 | 27 | 188 | 164 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| rpa3 |   | 2 | 53 | 194 | 180 | 0.011 | 0.107 | 0.109 | 0.001 | 0.162 | 0.191 | 0.126 | 0.132 | 0.149 | 0.043 |
|---|
| mcm2 |   | 4 | 27 | 188 | 164 | 0.151 | 0.254 | 0.188 | 0.128 | 0.154 | 0.267 | 0.224 | 0.200 | 0.101 | 0.012 |
|---|
| gtf2h4 |   | 1 | 90 | 194 | 195 | -0.158 | 0.147 | 0.020 | 0.115 | 0.112 | 0.190 | 0.187 | 0.053 | 0.006 | -0.039 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 2968-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.249E-07 | 2.873E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.059E-06 | 2.368E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.31E-06 | 1.771E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.841E-03 |
|---|
| thymus development | GO:0048538 |  | 8.466E-06 | 3.894E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.293E-05 | 4.955E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.832E-05 | 6.02E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 6.144E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 7.198E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.817E-05 | 6.478E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 2.817E-05 | 5.889E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.892E-08 | 9.507E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 6.393E-07 | 7.809E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 7.473E-07 | 6.085E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 1.487E-03 |
|---|
| thymus development | GO:0048538 |  | 4.782E-06 | 2.336E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 5.737E-06 | 2.336E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 9.122E-06 | 3.184E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.042E-05 | 3.183E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 1.042E-05 | 2.829E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.181E-05 | 2.886E-03 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.649E-05 | 3.663E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 4.652E-08 | 1.119E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.24E-07 | 8.709E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 8.486E-07 | 6.806E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.669E-03 |
|---|
| thymus development | GO:0048538 |  | 5.449E-06 | 2.622E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 6.537E-06 | 2.622E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.039E-05 | 3.573E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.188E-05 | 3.572E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 1.188E-05 | 3.175E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.346E-05 | 3.238E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 3.7E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.892E-08 | 9.507E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 6.393E-07 | 7.809E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 7.473E-07 | 6.085E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.436E-06 | 1.487E-03 |
|---|
| thymus development | GO:0048538 |  | 4.782E-06 | 2.336E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 5.737E-06 | 2.336E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 9.122E-06 | 3.184E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.042E-05 | 3.183E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 1.042E-05 | 2.829E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.181E-05 | 2.886E-03 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.649E-05 | 3.663E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 4.652E-08 | 1.119E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.24E-07 | 8.709E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 8.486E-07 | 6.806E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.669E-03 |
|---|
| thymus development | GO:0048538 |  | 5.449E-06 | 2.622E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 6.537E-06 | 2.622E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.039E-05 | 3.573E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.188E-05 | 3.572E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 1.188E-05 | 3.175E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.346E-05 | 3.238E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 3.7E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 4.652E-08 | 1.119E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.24E-07 | 8.709E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 8.486E-07 | 6.806E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.669E-03 |
|---|
| thymus development | GO:0048538 |  | 5.449E-06 | 2.622E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 6.537E-06 | 2.622E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.039E-05 | 3.573E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.188E-05 | 3.572E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 1.188E-05 | 3.175E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.346E-05 | 3.238E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 3.7E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.249E-07 | 2.873E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.059E-06 | 2.368E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.31E-06 | 1.771E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.841E-03 |
|---|
| thymus development | GO:0048538 |  | 8.466E-06 | 3.894E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.293E-05 | 4.955E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.832E-05 | 6.02E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 6.144E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 7.198E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.817E-05 | 6.478E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 2.817E-05 | 5.889E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.249E-07 | 2.873E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.059E-06 | 2.368E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.31E-06 | 1.771E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.841E-03 |
|---|
| thymus development | GO:0048538 |  | 8.466E-06 | 3.894E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.293E-05 | 4.955E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.832E-05 | 6.02E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 6.144E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 7.198E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.817E-05 | 6.478E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 2.817E-05 | 5.889E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.249E-07 | 2.873E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.059E-06 | 2.368E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.31E-06 | 1.771E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.841E-03 |
|---|
| thymus development | GO:0048538 |  | 8.466E-06 | 3.894E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.293E-05 | 4.955E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.832E-05 | 6.02E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 6.144E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 7.198E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.817E-05 | 6.478E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 2.817E-05 | 5.889E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.249E-07 | 2.873E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.059E-06 | 2.368E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.31E-06 | 1.771E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.941E-06 | 2.841E-03 |
|---|
| thymus development | GO:0048538 |  | 8.466E-06 | 3.894E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.293E-05 | 4.955E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 1.832E-05 | 6.02E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.137E-05 | 6.144E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.817E-05 | 7.198E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.817E-05 | 6.478E-03 |
|---|
| DNA replication initiation | GO:0006270 |  | 2.817E-05 | 5.889E-03 |
|---|



