Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2956-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1733 | 2.715e-02 | 1.256e-02 | 1.269e-01 |
|---|
| IPC-NIBC-129 | 0.1923 | 6.737e-02 | 5.620e-02 | 3.128e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1989 | 3.731e-02 | 2.847e-02 | 3.585e-01 |
|---|
| Loi_GPL570 | 0.2494 | 3.542e-02 | 4.827e-02 | 1.700e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1585 | 9.022e-03 | 4.281e-03 | 1.181e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3238 | 7.672e-02 | 3.875e-02 | 4.179e-01 |
|---|
| Schmidt | 0.2880 | 4.587e-02 | 4.809e-02 | 2.642e-01 |
|---|
| Sotiriou | 0.2316 | 1.237e-01 | 5.669e-02 | 6.376e-01 |
|---|
| Wang | 0.2240 | 3.153e-02 | 1.940e-02 | 1.260e-01 |
|---|
| Zhang | 0.2792 | 7.202e-03 | 3.528e-03 | 3.571e-02 |
|---|
Expression data for subnetwork 2956-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
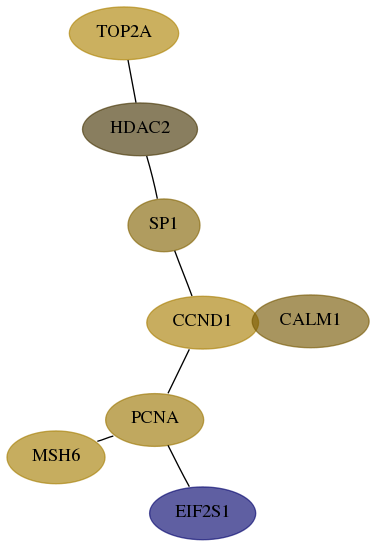
Score for each gene in subnetwork 2956-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| msh6 |   | 2 | 53 | 107 | 99 | 0.123 | 0.241 | 0.000 | -0.042 | 0.108 | 0.202 | 0.171 | 0.178 | 0.073 | 0.095 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| hdac2 |   | 16 | 10 | 64 | 39 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 |
|---|
| top2a |   | 22 | 7 | 70 | 46 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
GO Enrichment output for subnetwork 2956-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 6.218E-06 | 0.01430134 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 0.01427018 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 9.513E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.241E-05 | 7.135E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 8.106E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.86E-05 | 7.13E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.86E-05 | 6.112E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.985E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 2.602E-05 | 5.441E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 1.967E-06 | 4.806E-03 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 4.001E-06 | 4.887E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.001E-06 | 3.258E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.001E-06 | 2.443E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.016E-06 | 1.962E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.998E-06 | 2.442E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.998E-06 | 2.093E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 5.998E-06 | 1.832E-03 |
|---|
| response to UV | GO:0009411 |  | 6.074E-06 | 1.649E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 8.394E-06 | 2.051E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 8.394E-06 | 1.864E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 2.972E-06 | 7.151E-03 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 5.571E-06 | 6.702E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.571E-06 | 4.468E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.571E-06 | 3.351E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.571E-06 | 2.681E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.579E-06 | 2.638E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.353E-06 | 2.871E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 8.353E-06 | 2.512E-03 |
|---|
| response to UV | GO:0009411 |  | 8.889E-06 | 2.376E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.169E-05 | 2.812E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.169E-05 | 2.557E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 1.967E-06 | 4.806E-03 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 4.001E-06 | 4.887E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.001E-06 | 3.258E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.001E-06 | 2.443E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.016E-06 | 1.962E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.998E-06 | 2.442E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.998E-06 | 2.093E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 5.998E-06 | 1.832E-03 |
|---|
| response to UV | GO:0009411 |  | 6.074E-06 | 1.649E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 8.394E-06 | 2.051E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 8.394E-06 | 1.864E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 2.972E-06 | 7.151E-03 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 5.571E-06 | 6.702E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.571E-06 | 4.468E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.571E-06 | 3.351E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.571E-06 | 2.681E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.579E-06 | 2.638E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.353E-06 | 2.871E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 8.353E-06 | 2.512E-03 |
|---|
| response to UV | GO:0009411 |  | 8.889E-06 | 2.376E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.169E-05 | 2.812E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.169E-05 | 2.557E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 2.972E-06 | 7.151E-03 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 5.571E-06 | 6.702E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.571E-06 | 4.468E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.571E-06 | 3.351E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.571E-06 | 2.681E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.579E-06 | 2.638E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.353E-06 | 2.871E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 8.353E-06 | 2.512E-03 |
|---|
| response to UV | GO:0009411 |  | 8.889E-06 | 2.376E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.169E-05 | 2.812E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.169E-05 | 2.557E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 6.218E-06 | 0.01430134 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 0.01427018 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 9.513E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.241E-05 | 7.135E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 8.106E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.86E-05 | 7.13E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.86E-05 | 6.112E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.985E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 2.602E-05 | 5.441E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 6.218E-06 | 0.01430134 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 0.01427018 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 9.513E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.241E-05 | 7.135E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 8.106E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.86E-05 | 7.13E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.86E-05 | 6.112E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.985E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 2.602E-05 | 5.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 6.218E-06 | 0.01430134 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 0.01427018 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 9.513E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.241E-05 | 7.135E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 8.106E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.86E-05 | 7.13E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.86E-05 | 6.112E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.985E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 2.602E-05 | 5.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiosis I | GO:0007127 |  | 6.218E-06 | 0.01430134 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 0.01427018 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 9.513E-03 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.241E-05 | 7.135E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 8.106E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.86E-05 | 7.13E-03 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 1.86E-05 | 6.112E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.985E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 2.602E-05 | 5.441E-03 |
|---|



