Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2886-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1427 | 5.106e-03 | 1.286e-03 | 3.387e-02 |
|---|
| IPC-NIBC-129 | 0.1735 | 3.733e-02 | 2.867e-02 | 1.814e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2787 | 5.384e-02 | 4.359e-02 | 4.576e-01 |
|---|
| Loi_GPL570 | 0.2959 | 1.096e-01 | 1.363e-01 | 4.043e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1863 | 2.933e-02 | 2.048e-02 | 3.869e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3790 | 4.910e-02 | 2.186e-02 | 3.047e-01 |
|---|
| Schmidt | 0.2035 | 3.445e-02 | 3.635e-02 | 2.110e-01 |
|---|
| Sotiriou | 0.2499 | 7.932e-02 | 3.007e-02 | 4.947e-01 |
|---|
| Wang | 0.1704 | 7.941e-02 | 6.304e-02 | 2.550e-01 |
|---|
| Zhang | 0.3197 | 2.820e-03 | 1.240e-03 | 1.635e-02 |
|---|
Expression data for subnetwork 2886-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
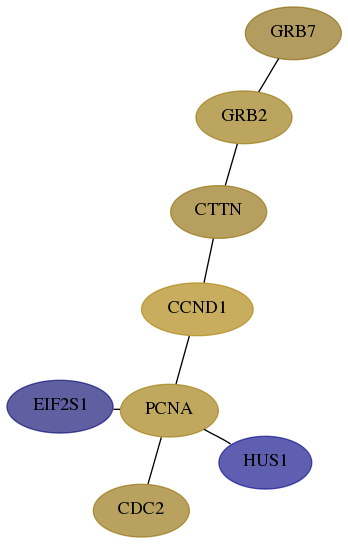
Score for each gene in subnetwork 2886-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| cttn |   | 5 | 20 | 8 | 9 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.216 | 0.069 | 0.115 | 0.129 | 0.025 |
|---|
| grb7 |   | 1 | 90 | 67 | 79 | 0.056 | 0.082 | 0.167 | 0.110 | 0.122 | 0.201 | 0.097 | 0.074 | -0.038 | 0.006 |
|---|
| hus1 |   | 6 | 17 | 22 | 17 | -0.058 | 0.197 | 0.116 | 0.109 | 0.101 | 0.160 | 0.072 | 0.082 | 0.081 | 0.071 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
| grb2 |   | 6 | 17 | 8 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 |
|---|
GO Enrichment output for subnetwork 2886-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.348E-06 | 5.4E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 0.01556281 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.54E-05 | 0.01180624 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.01205385 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 5.477E-05 | 0.02519411 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.587E-05 | 0.03291649 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 9.045E-05 | 0.0297204 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.03425095 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.191E-04 | 0.03044529 |
|---|
| response to light stimulus | GO:0009416 |  | 1.233E-04 | 0.02836839 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.078E-04 | 0.04345934 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.206E-07 | 1.028E-03 |
|---|
| response to UV | GO:0009411 |  | 3.338E-06 | 4.077E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.526E-06 | 2.871E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.565E-06 | 4.621E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.849E-05 | 9.036E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.104E-05 | 8.567E-03 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 2.831E-05 | 9.879E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.234E-05 | 9.876E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.114E-05 | 0.01388186 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 8.062E-05 | 0.01969543 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.731E-05 | 0.01939083 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.167E-07 | 1.484E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 5.88E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.777E-06 | 4.633E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.053E-05 | 6.336E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.929E-05 | 0.01409416 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.023E-05 | 0.01212194 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 3.94E-05 | 0.01354189 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.501E-05 | 0.01353679 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.407E-05 | 0.01712726 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 1.121E-04 | 0.02698206 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.121E-04 | 0.02452914 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.206E-07 | 1.028E-03 |
|---|
| response to UV | GO:0009411 |  | 3.338E-06 | 4.077E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.526E-06 | 2.871E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.565E-06 | 4.621E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.849E-05 | 9.036E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.104E-05 | 8.567E-03 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 2.831E-05 | 9.879E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.234E-05 | 9.876E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.114E-05 | 0.01388186 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 8.062E-05 | 0.01969543 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.731E-05 | 0.01939083 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.167E-07 | 1.484E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 5.88E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.777E-06 | 4.633E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.053E-05 | 6.336E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.929E-05 | 0.01409416 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.023E-05 | 0.01212194 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 3.94E-05 | 0.01354189 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.501E-05 | 0.01353679 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.407E-05 | 0.01712726 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 1.121E-04 | 0.02698206 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.121E-04 | 0.02452914 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.167E-07 | 1.484E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 5.88E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.777E-06 | 4.633E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.053E-05 | 6.336E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.929E-05 | 0.01409416 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.023E-05 | 0.01212194 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 3.94E-05 | 0.01354189 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.501E-05 | 0.01353679 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.407E-05 | 0.01712726 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 1.121E-04 | 0.02698206 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.121E-04 | 0.02452914 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.348E-06 | 5.4E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 0.01556281 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.54E-05 | 0.01180624 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.01205385 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 5.477E-05 | 0.02519411 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.587E-05 | 0.03291649 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 9.045E-05 | 0.0297204 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.03425095 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.191E-04 | 0.03044529 |
|---|
| response to light stimulus | GO:0009416 |  | 1.233E-04 | 0.02836839 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.078E-04 | 0.04345934 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.348E-06 | 5.4E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 0.01556281 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.54E-05 | 0.01180624 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.01205385 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 5.477E-05 | 0.02519411 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.587E-05 | 0.03291649 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 9.045E-05 | 0.0297204 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.03425095 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.191E-04 | 0.03044529 |
|---|
| response to light stimulus | GO:0009416 |  | 1.233E-04 | 0.02836839 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.078E-04 | 0.04345934 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.348E-06 | 5.4E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 0.01556281 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.54E-05 | 0.01180624 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.01205385 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 5.477E-05 | 0.02519411 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.587E-05 | 0.03291649 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 9.045E-05 | 0.0297204 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.03425095 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.191E-04 | 0.03044529 |
|---|
| response to light stimulus | GO:0009416 |  | 1.233E-04 | 0.02836839 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.078E-04 | 0.04345934 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.348E-06 | 5.4E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 0.01556281 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.54E-05 | 0.01180624 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.01205385 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 5.477E-05 | 0.02519411 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.587E-05 | 0.03291649 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 9.045E-05 | 0.0297204 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.03425095 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.191E-04 | 0.03044529 |
|---|
| response to light stimulus | GO:0009416 |  | 1.233E-04 | 0.02836839 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.078E-04 | 0.04345934 |
|---|



